

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
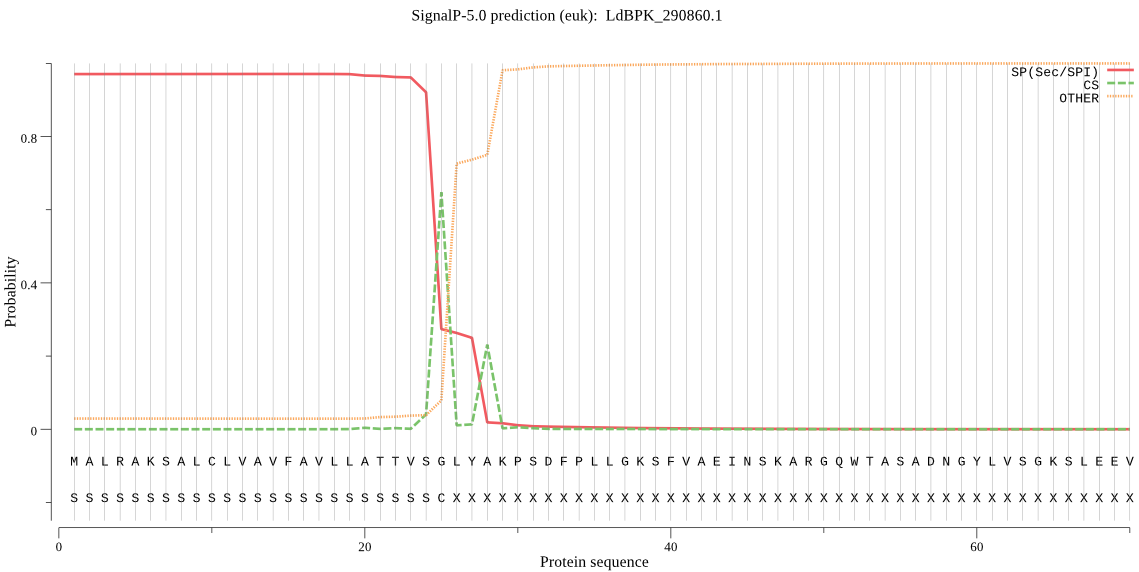
| LdBPK_290860.1 | SP | 0.004440 | 0.995498 | 0.000062 | CS pos: 25-26. VSG-LY. Pr: 0.6103 |
MALRAKSALCLVAVFAVLLATTVSGLYAKPSDFPLLGKSFVAEINSKARGQWTASADNGY LVSGKSLEEVRKLMGVTDMSTEAVPPRNFSVDEMQQDLPEFFDAAEHWPMCVTISEIRDQ SNCGSCWAIAAVEAISDRYCTLGGVPDRRISTSNLLSCCFICGFGCYGGIPTMAWLWWVW VGITTEVCQPYPFGPCSHHGNSDKYPPCPNTIYDTPKCNTTCEKSEMDLVKYKGGTSYSV KGEKELMIELMTNGPLEVTMQVYSDFVGYKSGVYKHVSGDLLGGHAVKLVGWGTQGGVPY WKIANSWNTDWGDKGYFLIQRGSNECGIESGGVAGTPAQE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_290860.1 | 225 S | TCEKSEMDL | 0.995 | unsp | LdBPK_290860.1 | 225 S | TCEKSEMDL | 0.995 | unsp | LdBPK_290860.1 | 225 S | TCEKSEMDL | 0.995 | unsp | LdBPK_290860.1 | 66 S | VSGKSLEEV | 0.992 | unsp | LdBPK_290860.1 | 151 S | DRRISTSNL | 0.997 | unsp |