

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_291680.1 | OTHER | 0.999232 | 0.000517 | 0.000251 |
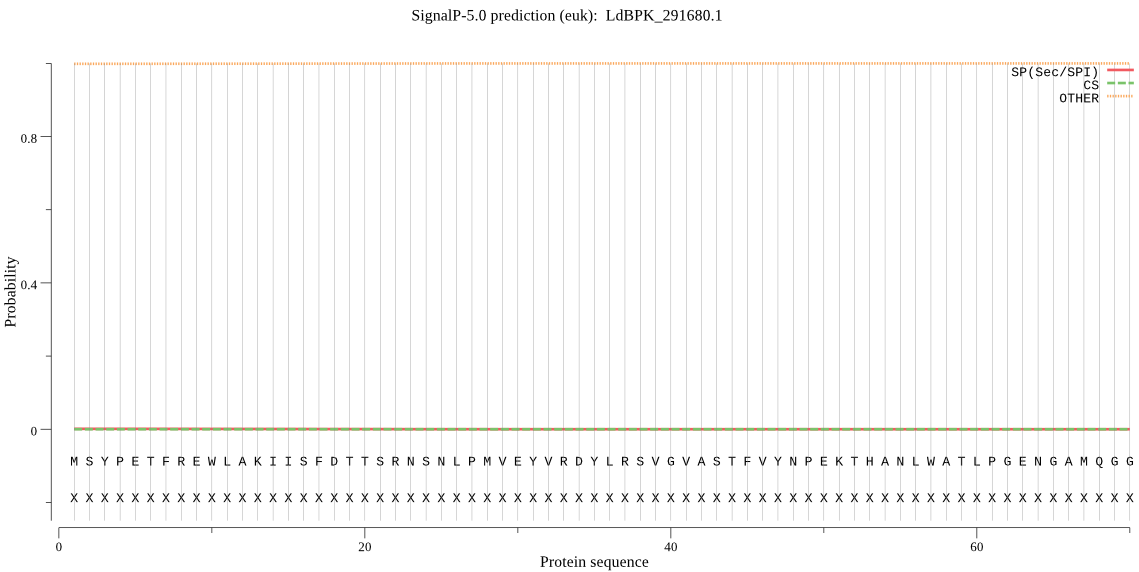
MSYPETFREWLAKIISFDTTSRNSNLPMVEYVRDYLRSVGVASTFVYNPEKTHANLWATL PGENGAMQGGIVLSGHTDVVPVDGQKWDSDPFTMVEKDGKLFGRGACDMKGFLAVVLALT PQFLRMNRVKPVHYAFSFDEEVGCTGVPYLIEYLKAHGFLADACLIGEPTDMNVYVGSKG FTQWSVSVQGKAIHSSMALMNTSCNAIEYAAQIITKVREIAVDLRKNGRQDPEYACPFPC ITTGLIKGGNAVNTVPAQCEFVVTARITDNETPDAIERRVRSYVNDHVLPAMREEYPDAE VKVTRPLSMPAFNGKEAASVTQKALMLRKATRTYKTGGGTEGGYFEDVLGVSTVIVGPGP QAMAHLPNEYVLVDQMEKSTEFTMALVRLYTDPSYCGAGNL