

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_292470.1 | OTHER | 0.999077 | 0.000233 | 0.000690 |
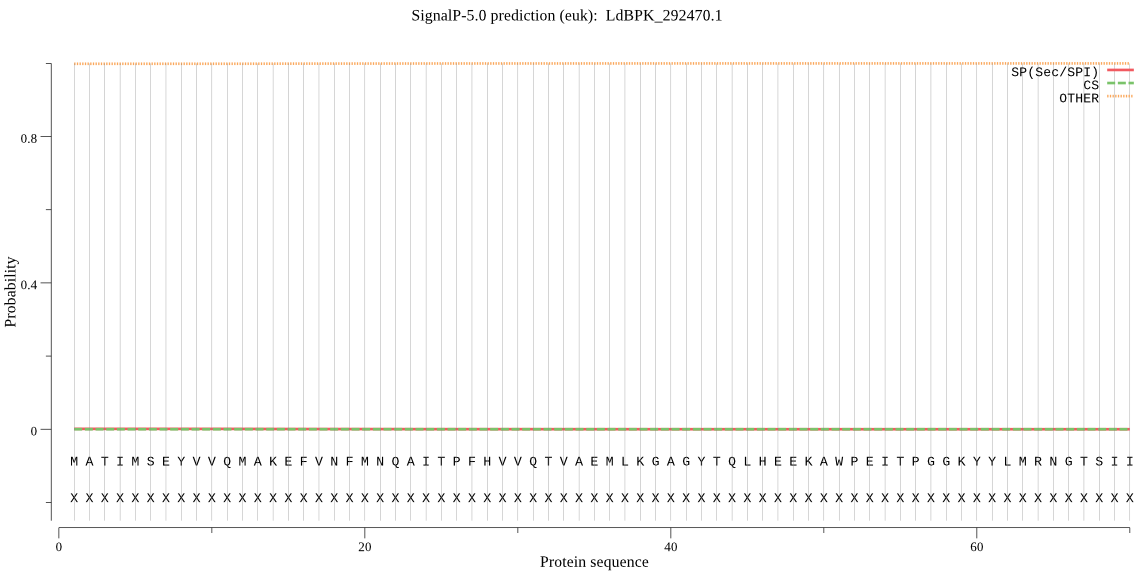
MATIMSEYVVQMAKEFVNFMNQAITPFHVVQTVAEMLKGAGYTQLHEEKAWPEITPGGKY YLMRNGTSIIAFSVGGKFDPMNGVKIVGAHTDSPNFLLKPRTKSTAADYQRVAVQCYGGG LWHSWFDRDLTVAGRVMISRERLEQKIIKIDKPIMRIPNLAIHLTSAKDREAFSPNKESH LIPIISTQIAAKIAECDDKDASNPNHCVSLMKAIASVAGCSADEIVDFDLSVIDTQPAVI GGIHDEFIFSPRLDNLISCYCAVKAIVEAGSLENDTMMRMVCLFDHEECGSSSSQGAAGS LVPDVIEHIVSNKTLRATLVANSFLLSVDGAHGCHPNYADKHENAHRPALHGGPVIKYNA NVRYATNGLTAAVVKDMAKKADIPIQEFVVRNDSPCGSTIGPILSSLSGIKTADIGNPMI SMHSIREMCGTVDVYYLTKLIESFFVNYQSPHE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_292470.1 | 424 S | ISMHSIREM | 0.994 | unsp | LdBPK_292470.1 | 424 S | ISMHSIREM | 0.994 | unsp | LdBPK_292470.1 | 424 S | ISMHSIREM | 0.994 | unsp | LdBPK_292470.1 | 166 S | IHLTSAKDR | 0.995 | unsp | LdBPK_292470.1 | 394 S | VRNDSPCGS | 0.993 | unsp |