

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_292880.1 | OTHER | 0.909908 | 0.074110 | 0.015982 |
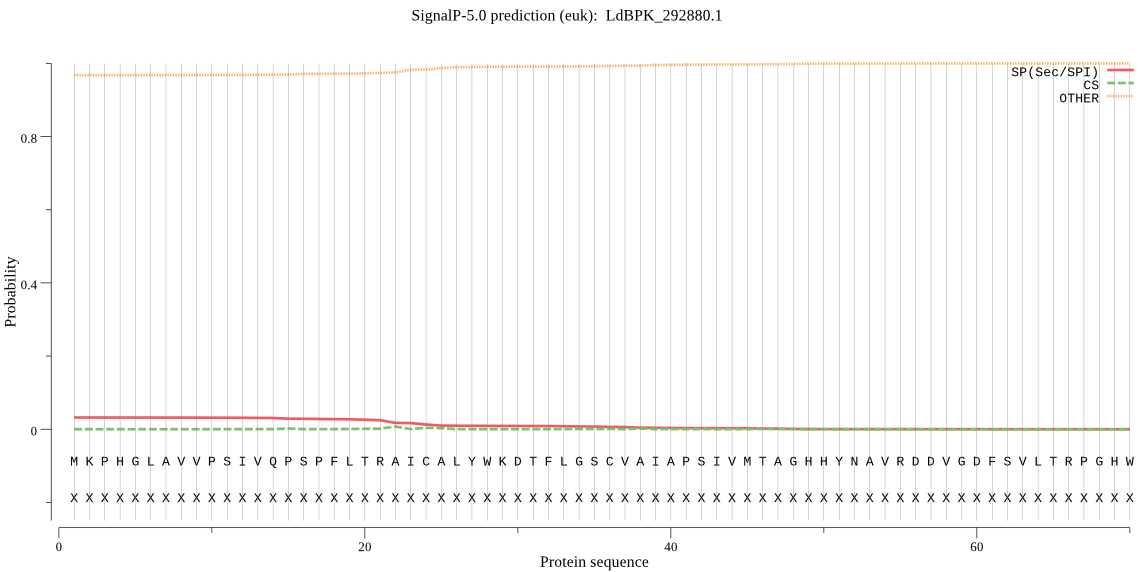
MKPHGLAVVPSIVQPSPFLTRAICALYWKDTFLGSCVAIAPSIVMTAGHHYNAVRDDVGD FSVLTRPGHWTAVEYASKDSTYDVVVFWLCEEVTTHTSLRGYLPAVGAQVATVWLSPKPP HDAIVSPGVVMESEAEKCVARGTVSMTGSSGAPVVDVFGEHIVGLHLTSNTRDGSRVSGF IPSRKLVSLFAEMWVEHARVLSSA
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LdBPK_292880.1 | 80 S | ASKDSTYDV | 0.992 | unsp |