

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
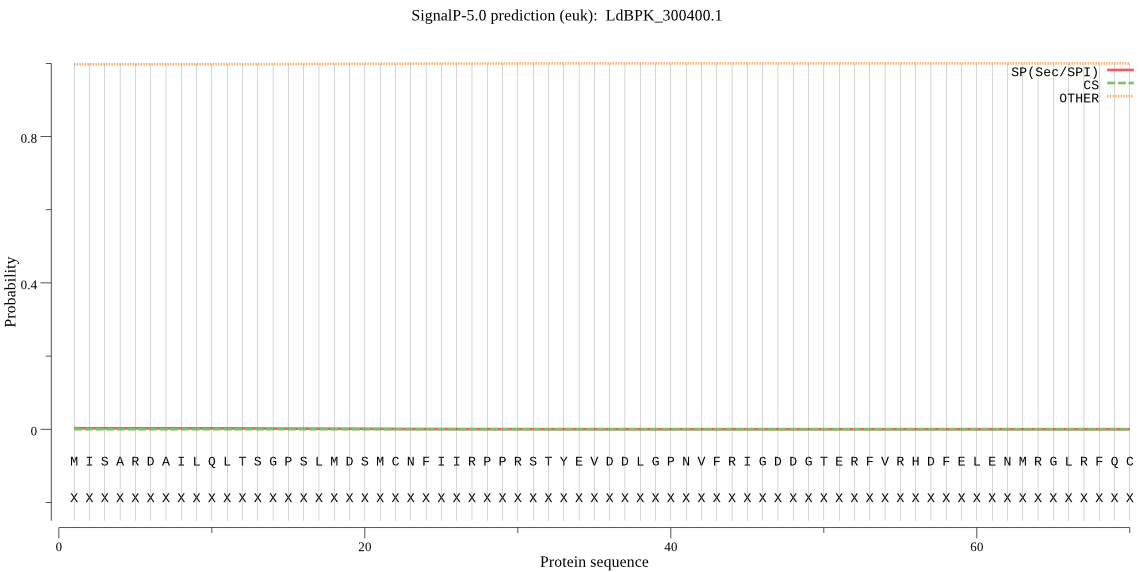
| LdBPK_300400.1 | OTHER | 0.970881 | 0.009950 | 0.019169 |
MISARDAILQLTSGPSLMDSMCNFIIRPPRSTYEVDDLGPNVFRIGDDGTERFVRHDFEL ENMRGLRFQCSWFKTYPARRVPCVVYCHANCGGRYDGLEALFLLREGFSLFCFDFCGSGM SEGEYISLGFYERQDLVAVVEFLTLKSDEVDGVALWGRSMGAVAAIMYASKDPWIRCIVC DSPFASLRLLIDDLVERHGGRTARVLPKILVHGIVERIRKRIMKRAAFDIDDLDAVKYAK ACGVPALLFHGADDDFVSPTHCEMIRDAFPIPCLQQFTPGGHNCDRQEDIHVLIRAFLRL YLIDKPQGAREIQAARELQLANTASPAAVAAAAANKGAISRLSAPAALAPKQLVPPASYG MRCAETALISSSSTSATASSSTTSPSTSASSSTSFDSSLPLPAALSVPLSPENRLVYDCS NSRDSTPPPKAAATLASSLPPSLTPFRSSSGTRAHVSASTATVAGEGHATSGATVPPPSS PAADSTDGLQNCTASP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_300400.1 | 384 S | SSTTSPSTS | 0.99 | unsp | LdBPK_300400.1 | 384 S | SSTTSPSTS | 0.99 | unsp | LdBPK_300400.1 | 384 S | SSTTSPSTS | 0.99 | unsp | LdBPK_300400.1 | 410 S | SVPLSPENR | 0.99 | unsp | LdBPK_300400.1 | 425 S | NSRDSTPPP | 0.997 | unsp | LdBPK_300400.1 | 31 S | RPPRSTYEV | 0.995 | unsp | LdBPK_300400.1 | 125 Y | SEGEYISLG | 0.991 | unsp |