

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_311150.1 | OTHER | 0.999051 | 0.000486 | 0.000463 |
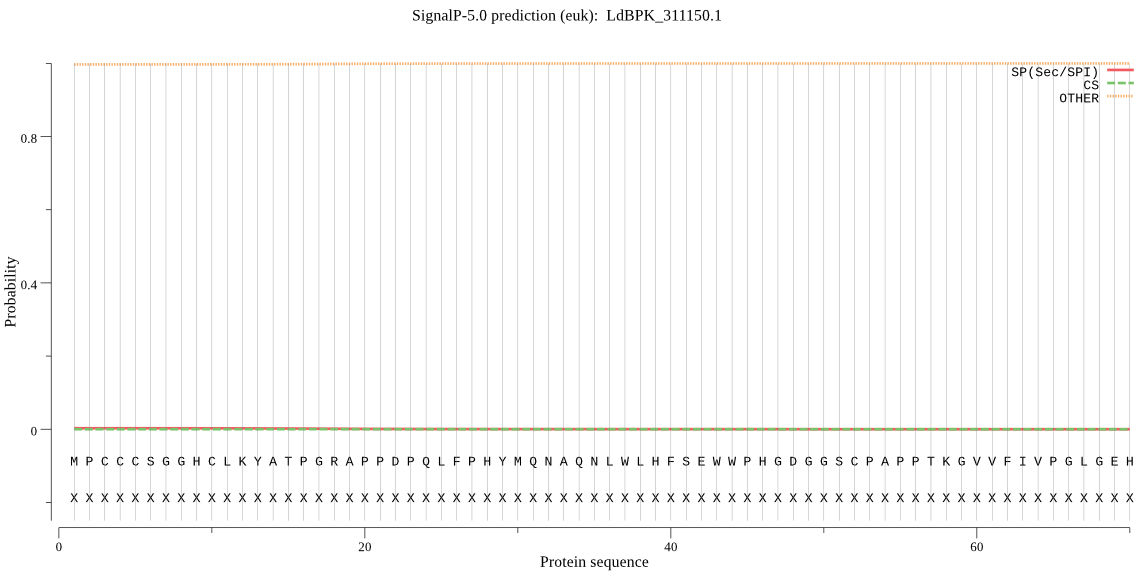
MPCCCSGGHCLKYATPGRAPPDPQLFPHYMQNAQNLWLHFSEWWPHGDGGSCPAPPTKGV VFIVPGLGEHTGRYDSVALRLNQEGYVVFSMDNQGTGGSEGERLYVERFTHFVDDVCAFV VFIQTRYAALKSQPTFLMGHSMGGLIATLVAQRDASGFRGVVLSGPALGLSKPVPRFMRS LAHFLSQWFPKLPVRKLNPELVSYNTPVVQLVKQDPFYSNVTLRARFVDEMLIAQDRAAE AAATSKFPFLIVHGEEDQLCSLETSKSFFKNALSEDKFLASYRRAGHEVLTELCRAEVMA EVMKFINERAR
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LdBPK_311150.1 | 156 S | QRDASGFRG | 0.993 | unsp |