

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_312040.1 | SP | 0.100618 | 0.892803 | 0.006579 | CS pos: 28-29. TVA-AP. Pr: 0.5410 |
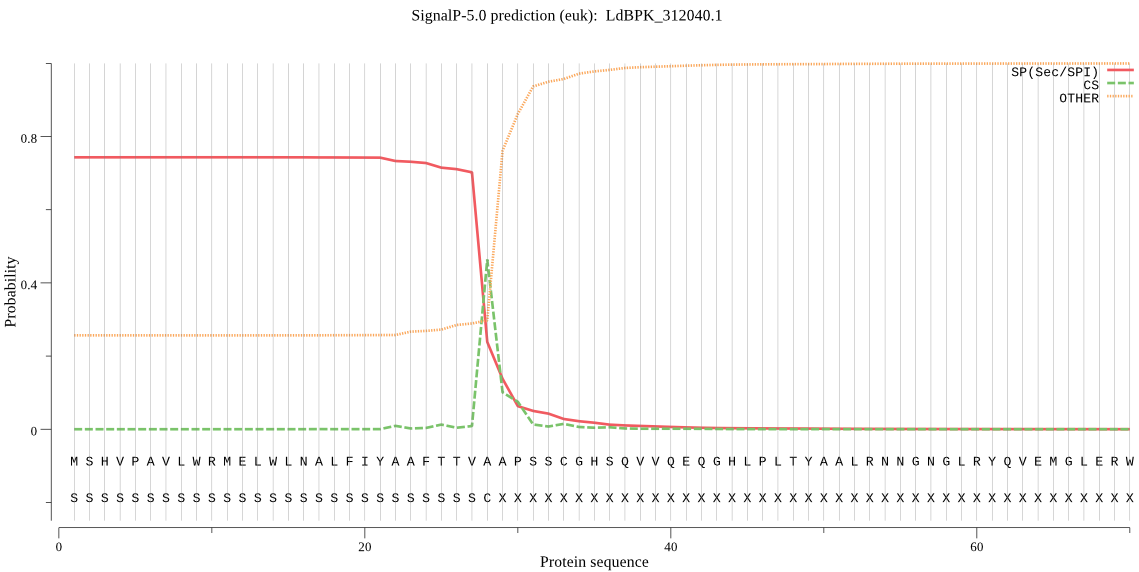
MSHVPAVLWRMELWLNALFIYAAFTTVAAPSSCGHSQVVQEQGHLPLTYAALRNNGNGLR YQVEMGLERWNGSATAEQHAVEPQATVTISSGREDSEWAPIRIAVFMPDLEDESKYCTAV GQMRPDSSGSLVPCTSNADLLTDAKKAVLINSILPQAVNMHAQRLKVRRLAASTSIVVNS MPGSYCGSFTVPTAHRTTGVPNADFVVYVAAGPTSTPKSFIAWAVTCQYYPNTATITSRP AVGAMYFNPRYLPTSVGETQEHIDRYGGNPSGSTNRLRRVAAHELLHALGFTSSVFKARG MFATVPSLRGKRNVPVLNSSSVRAAAKAKYGISDAEVFYGAELEDQGAPGTSLSHWKRRA AKDELMAPVLSLARYSSLSLAALEDMGFYKVDFSKAEPVALGATAGGKLFTEPCLTEGTS NTPTVFCDSLSPSVRSCTADRLSIGRCALTTYSSSLPSYAQYFPNQPTLGGSLSHSDYCP VIQPLSNTGCSGGSLGAMPGSITGDNARCFDADNLLVKFTFTQPGAICAKVHCKSASRTY EVLVIGANSWLSCGAGGTGVTVQPAVSSPNVFVEGGLIACPPYDDVCYANPVAFAEVEAA PTTSTTASPSAATAATHGMVVSGLVAVAVLLGSGIC
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_312040.1 | 91 S | VTISSGRED | 0.994 | unsp | LdBPK_312040.1 | 255 S | YLPTSVGET | 0.996 | unsp |