

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
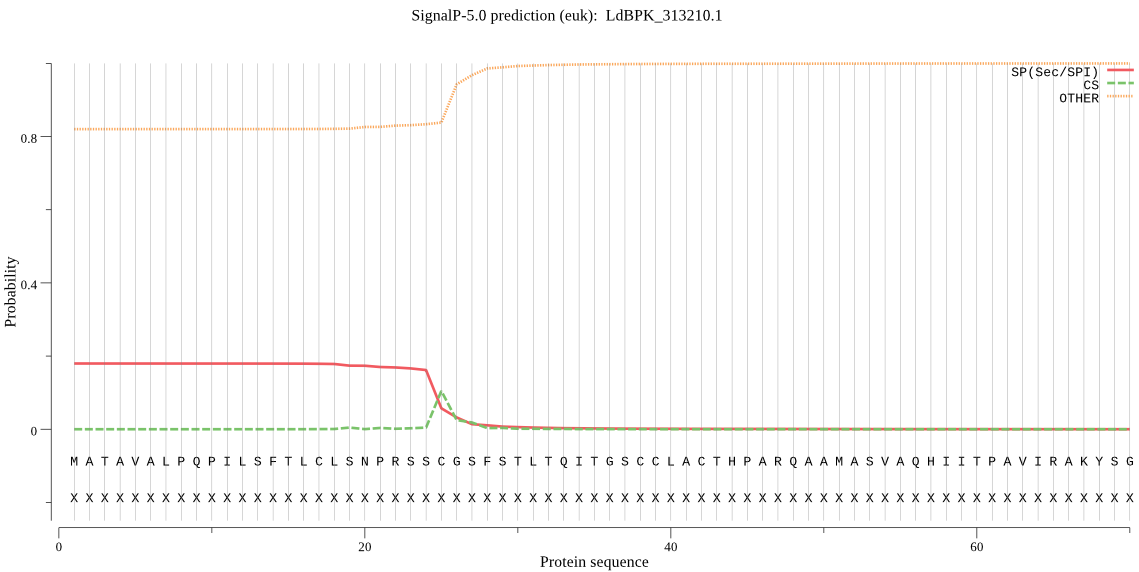
| LdBPK_313210.1 | SP | 0.216627 | 0.613326 | 0.170047 | CS pos: 25-26. SSC-GS. Pr: 0.5336 |
MATAVALPQPILSFTLCLSNPRSSCGSFSTLTQITGSCCLACTHPARQAAMASVAQHIIT PAVIRAKYSGYVLSNGVKCVIVQDANAKMPAAAMCIRAGQLNDPVELPGLAHFCEHMLFM GTEKFPKEDEFDSFVSKASGLTNAFTEDCDTVYYFSVSDGSLEGALERFVEFFASPSFSA GAVAREVNAVHSEDEKNHNSDYWRLDELIRDFCNPKHPRSRYGNGNLTTLRDEPQRRGID VRESLKTFHSRYYLADGATIVVVSMRSADEVLSLIEGPLARMKQGAVPRFSFLEAGEHLF TSAALGSWTNVRTVQKMRELRLMWAVRSPSSAWRTTPSAYIAHLLGHECDTSVLGVLKKR NWATGMLAGSHRVDDDFEVLDASVTLTVEGFRNALKVVDLLYQGIGQSIAHGVDAEVYKG MKAEERLFFESVDAGSPANHCVALAQNANATDLEHCWIGGNVVLEDDMEAVLAYVRQLTP QNCVMVMQWGDMPIDAATGATPAEGNPVESHAQQHDEEDDEDSQSAEEETAEETEQASES NASAEELFLSLPAFAQVRVSNVSRFHRALYQTVKIPEEHEARWQSLISGPYPPELALPST NPFIATDFTIYPPAAEEAVDEIASPHAVTYVRKDAGHHSTFKMALRCNVLSPVASASPLN RLYTRVMHGILSNALTEMAYYAMLASLTNEVIFSPTGLGFAVEGPSQKLYEFFFAVVRKG LSVQVLQGTAEEYATYLETGVQKLKNVGMAQPYKVLHETQKKATRHTYYLFNEMIACESA ATYEGYCTFVKQYLESGLLLECFVAGNVLSTQDVREMLVGRLEDLLGTLTIPIPLKETIP RVRDTYGPRAGDGKANESVLCSFDVLSMPPSNPADPNAAVVVDVIIGEATPRVMALTDTA MKLISSSFFNVLRTREALGYVVFAESSVDYCTAHALFVVQSALKDVDCVYLLSRIVAFLS AVEAQLDTVCSAEEVEKVKRGLIAALEKVPDSVEGDVKRLEGEYVSLSKFESRQRTIAAL LAITAEDVKSHLRNYFFNNRSESRALALCINNARTVAADVLAEPGNRRVPLPSVRSRQED SDDAPGDASEDTEPLTLPTFSDGSSCVEITSYASALEYQHGLCLVKCNAY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_313210.1 | 264 S | IVVVSMRSA | 0.992 | unsp | LdBPK_313210.1 | 264 S | IVVVSMRSA | 0.992 | unsp | LdBPK_313210.1 | 264 S | IVVVSMRSA | 0.992 | unsp | LdBPK_313210.1 | 525 S | EDSQSAEEE | 0.996 | unsp | LdBPK_313210.1 | 543 S | ESNASAEEL | 0.996 | unsp | LdBPK_313210.1 | 992 S | KVPDSVEGD | 0.996 | unsp | LdBPK_313210.1 | 156 S | VYYFSVSDG | 0.991 | unsp | LdBPK_313210.1 | 244 S | DVRESLKTF | 0.995 | unsp |