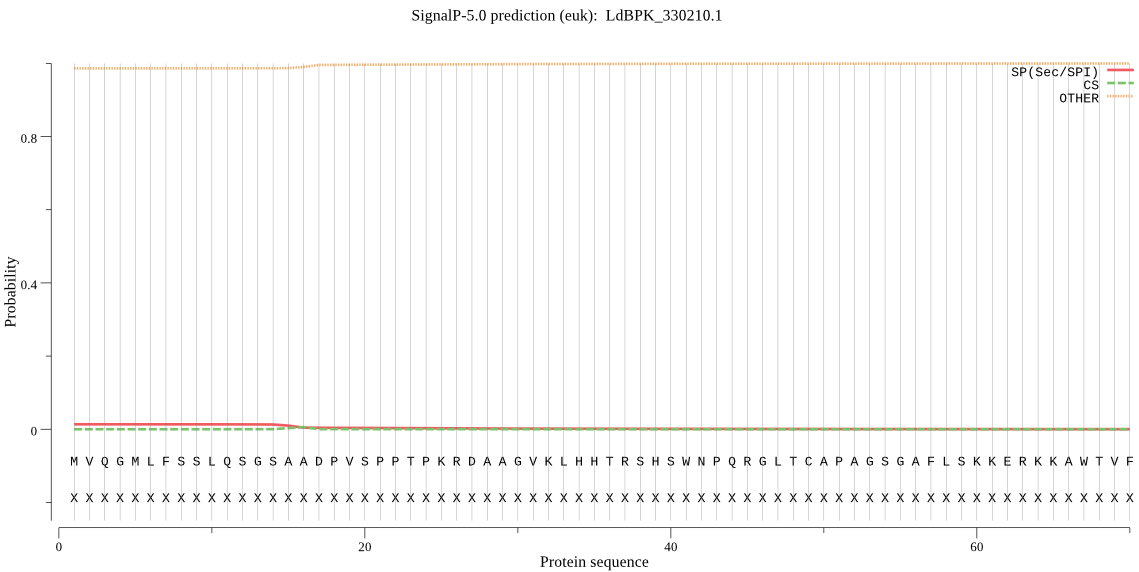
Fasta :-
MVQGMLFSSLQSGSAADPVSPPTPKRDAAGVKLHHTRSHSWNPQRGLTCAPAGSGAFLSK
KERKKAWTVFLHKNTHAFALIVNACRSSLAMRELQVGSVLLTFLYNVLRHMPGDKQRKAV
ALCEKLGIIRVCTSVLQEGKRRLRPGDVDAAFVASLRACEAAALLFTLGSTFNVKLWTLF
RLSQSLECTFACASVTFRLLEAEFARYHASLQSDLCQTTTAAALQEYTRKHTRKETLTTL
RRFQSRTQWTCTAFSHFCFTLFAFTGSSASAVDLGARGTELCATAIAMSTFFLTQIAPLL
LMSVKYDASEKTRHPDVGALVSIDAEHWVPSWVMRLMRRVEFMLLWCVALMQRLTGFDKV
RQMAAEVALRFHVPRGIVFYLLPPSTHMLQILPRRECVHVILLLLNALLAADREAVLADI
KDLGGLKVVVASVLALPEKSSQQWHHFYLQLCSMYGYTMLPSWNGRPACYVDERLPASLR
RQFDVARTPGGKSHLSEPVGSCHSAVDTRADASQARPSSTQAPLAQCAGRSTETVLDSII
SPSLTAARSGRGGSDNCLLLVDIRPDVFSPELRSGDVDPVSELVAFDPSQLPWPFKDAAP
PTTAESWYGASSSVAAAAEDAHLSVPLINVCEEDRVRVLRHHIARLTLLDEAACDGGPTP
HVHKVVFERLSSASPGAAGDAPSAVQGSADGVEFFSDYESGNLQRAIEVTDSEYDLVLSW
DTATNSYTQWFSFGMRHFTPGKTYRFNILNMEKGDSTFNEGQKPLLLHVPDGGNSSDPSA
PRPKWTRAGEGIFYFGNAFLRPARAHYFGKTGEAASAPAASPLSMSPRPLSMSVSPCAPV
SHLGGRAPPPVIAKKRIPALPTLSPPPAMPPPFTASTPAESGVLKQKCYFTVTFSVTMPT
TVAGTVYLANCFPFTYTEMRDHLAWLARESHACPTRSLLPQCLCWTPGGLQVPLLTVTAL
RNRDTGEPYTADEIRRRPIALLVARVHPGETNASWVMQGLLDTLLCPPAVAAEYASHLCD
NFVFKVIPMLNADGVIMGNHRCSFAGADLNRDYTNPKADFNPTLYAMKQLLRYWTGEEKR
QVVMFADFHGHSRAKNFLVYGCTMNTIRGVATHSRHKGTRGGDGGRRRRTRRASSTAPAG
PEKVIAALLSQLFPSFSLSQSSFAVTKDKKGSARVVLYDEFGVRMSYGFEATMVGGLVCV
PELLRVSAGTHDDDAAAPVSAVCKAEVHYSPTVFRAMGDVFLRALSILFRQWAALTALAA
DSGCSDPKSSQSDVLKSAALQLSWSMTAAAGGGREPPKNPFSPSSRESGAALPRAGQYAA
ATREDRRVGSPVRARSPAAPNEACSGLSVVPVAGQVEALDYLFMADCREGATPAKDEDGK
DEEGSIASGSDISTEEDSDDAEMEEDELLGTTSARHTRRRRSEAQYAGNASEVSEHSASS
PVGSGSDDQDWSGCNSSGSAADDSDDSRSSVSSLDEEIPTNLFT