

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
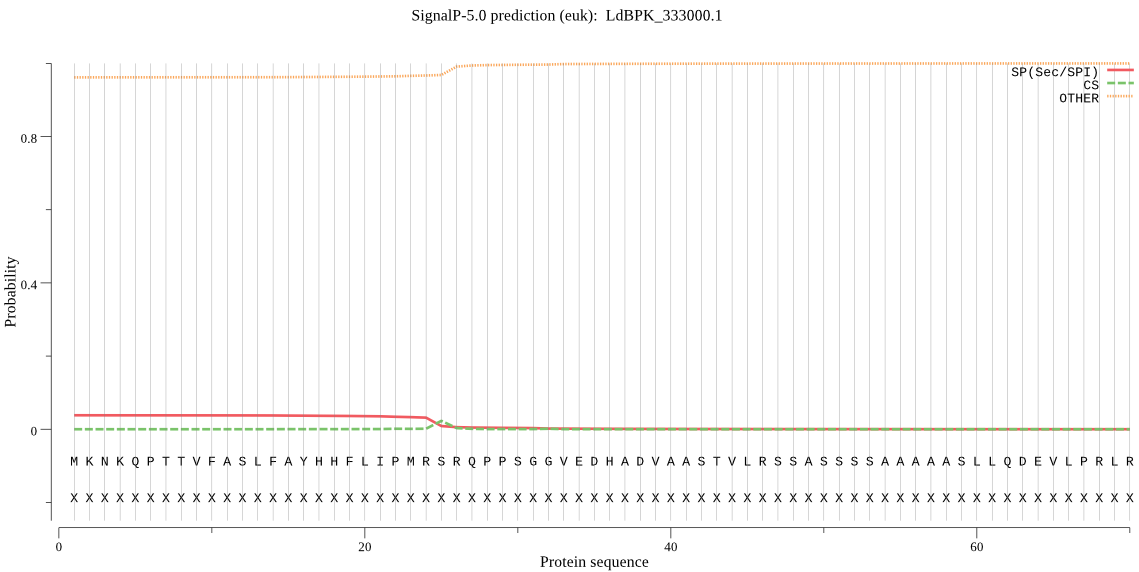
| LdBPK_333000.1 | OTHER | 0.910507 | 0.085823 | 0.003671 |
MKNKQPTTVFASLFAYHHFLIPMRSRQPPSGGVEDHADVAASTVLRSSASSSSAAAAASL LQDEVLPRLRKESGPSNAAICDAYATRLRQEATEEERRRGPHWRARCSPSILVALLIWAT IALVVYMKVDNSRLRPGAAPDTTSVETPLSQLPESCRGHYCLREGGKEPFGAVLGAHDGV FAYSNCNSDTCTSLLKYQMAIPLPPGARTALDAPHATTRLMTTGMKWQCVEFARRYWMLR GKPTPAFFGPVVGAADIWDTLTHVTFLDNATTAPLLKFKNGARLGYGGSAPRVGDLLIYP RDAEGFFSYGHVAVVVRVEMTTKAEADDSYMDAAVTSSKPRQRHGLVYLAEQNWDSATWP NPYHNYSRSLPLVVLESAEGLPLQYTIEDSIHGIQGWVRYDDEP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_333000.1 | 329 S | EADDSYMDA | 0.995 | unsp | LdBPK_333000.1 | 329 S | EADDSYMDA | 0.995 | unsp | LdBPK_333000.1 | 329 S | EADDSYMDA | 0.995 | unsp | LdBPK_333000.1 | 48 S | VLRSSASSS | 0.995 | unsp | LdBPK_333000.1 | 51 S | SSASSSSAA | 0.991 | unsp |