

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_333280.1 | OTHER | 0.892365 | 0.085321 | 0.022314 |
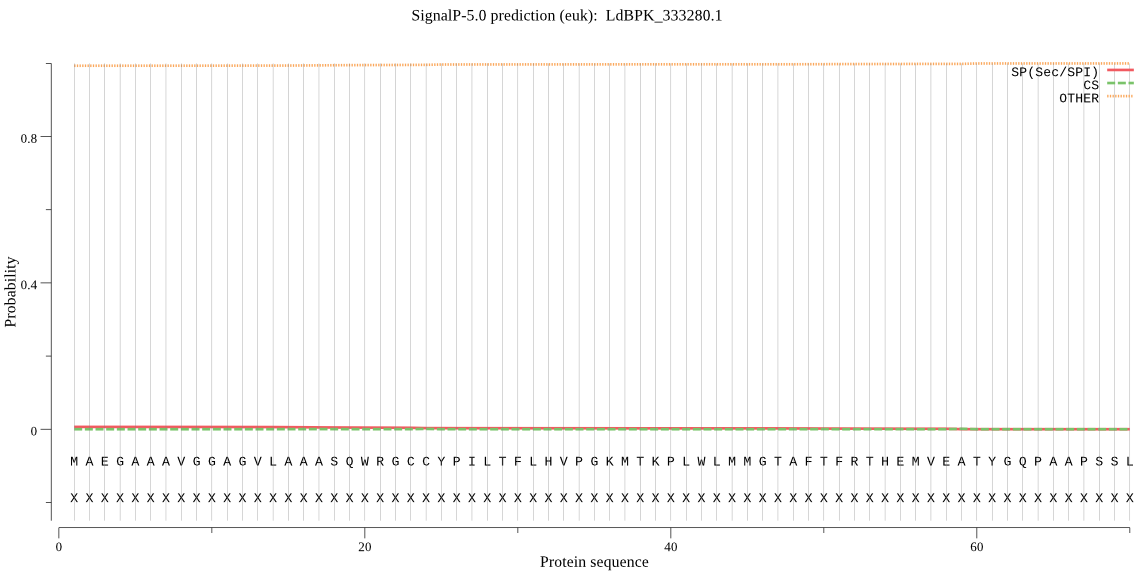
MAEGAAAVGGAGVLAAASQWRGCCYPILTFLHVPGKMTKPLWLMMGTAFTFRTHEMVEAT YGQPAAPSSLLLTAKHSFSPWDYTKDASQLKIPKEYQKPRFVIGRVYRTDADGRAVAADA VGLRLLAHHPIHDVALLAVDAASLKLLQCATYDSPAHALDLAHEALPLCKEPYPAASDGL LIGFRGAGRLGELDTLDPSVLQRLPPHEREALIKDLQDVEGKQRRATTTVSVLDARGMCK GVGDLATCYHGMSGGPLVTTNGQCAGVLYGHHPDAPGCIGYTPCADFAGWLEEEVQRYRK RCDLH
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LdBPK_333280.1 | 79 S | KHSFSPWDY | 0.994 | unsp |