

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
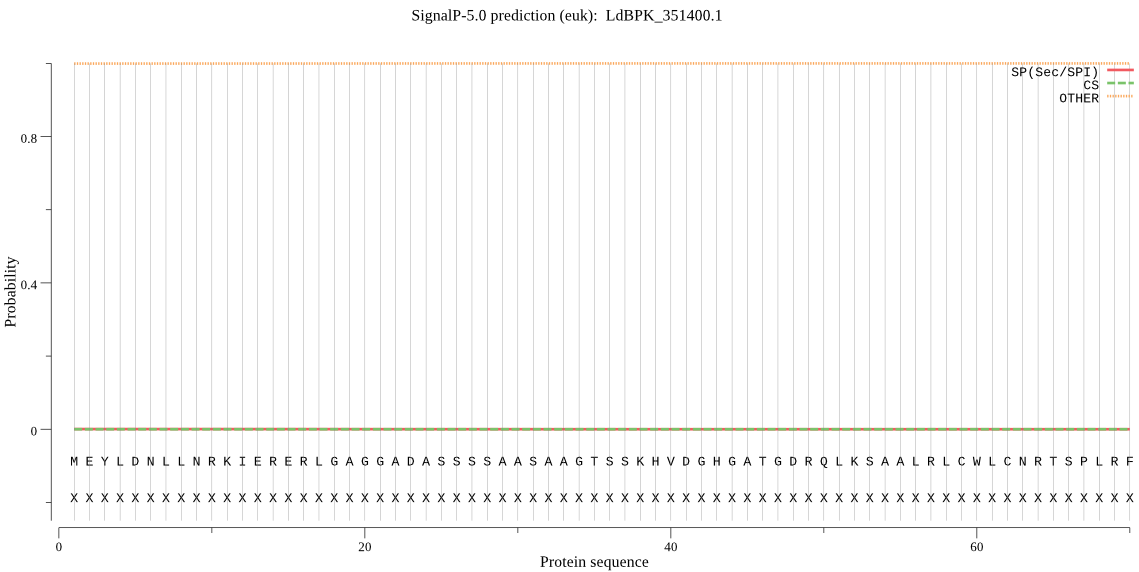
| LdBPK_351400.1 | OTHER | 0.999986 | 0.000001 | 0.000013 |
MEYLDNLLNRKIERERLGAGGADASSSSAASAAGTSSKHVDGHGATGDRQLKSAALRLCW LCNRTSPLRFRLQGGIHLCFTCFRASYLSLLLPIINTPARRASDARFLKLIGSIENERAG SVSSPTDELLASMYGEVIEPQQASFVMSSSMMRLRREEEEQRLLSATRRPRSGGAASSAK SVKSFWNCPLCTFLNAAVARECEACGFVNPGEVTCPVCKARCSLGMIGTPLSEENPASRC KTGEPHCIWNCRECGGLNTLDGDRCTSCRQPRYWACSQCTALHHMARGEDGLRYCPTCGA YNTPDDILVGQAKIDKETRYAGMVTAQQSAAQRRKSGSKDHDHNRGGSRAAAGGAAAAKA DDQIVFGVNDAHTLEELERQKQIENNEKRLLSRLNRLHISRNLQKTDGNCLFRALANQLF GQPRLHYLVRSLATAYMSEHSEDYAILFDGPAEWKKYLTAMKEQGTWGDELCLNAAARCF RVNIHVITSDQERWHIVFQHDQLGRTRTVRSDEDARTNNMAQTLTSYEGVSLFLAYLSPV HYDDITPLPVAHLQLGELLSGELNKRMKRNQTTHSPSDSGSSTSALASAGPSSGGRWADA SNFSHLPSRPLLADPAKPRTRSSDDLVKSCLESPTVSNATAPPVPRSRPHSCKPDTYKSS EPPSRPPPTSPTSLPSEYTC
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_351400.1 | 172 S | RRPRSGGAA | 0.996 | unsp | LdBPK_351400.1 | 172 S | RRPRSGGAA | 0.996 | unsp | LdBPK_351400.1 | 172 S | RRPRSGGAA | 0.996 | unsp | LdBPK_351400.1 | 181 S | SSAKSVKSF | 0.991 | unsp | LdBPK_351400.1 | 336 S | QRRKSGSKD | 0.997 | unsp | LdBPK_351400.1 | 511 S | RTVRSDEDA | 0.998 | unsp | LdBPK_351400.1 | 575 S | QTTHSPSDS | 0.997 | unsp | LdBPK_351400.1 | 622 S | PRTRSSDDL | 0.996 | unsp | LdBPK_351400.1 | 651 S | SRPHSCKPD | 0.998 | unsp | LdBPK_351400.1 | 670 S | PPPTSPTSL | 0.992 | unsp | LdBPK_351400.1 | 103 S | ARRASDARF | 0.991 | unsp | LdBPK_351400.1 | 124 S | GSVSSPTDE | 0.996 | unsp |