

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
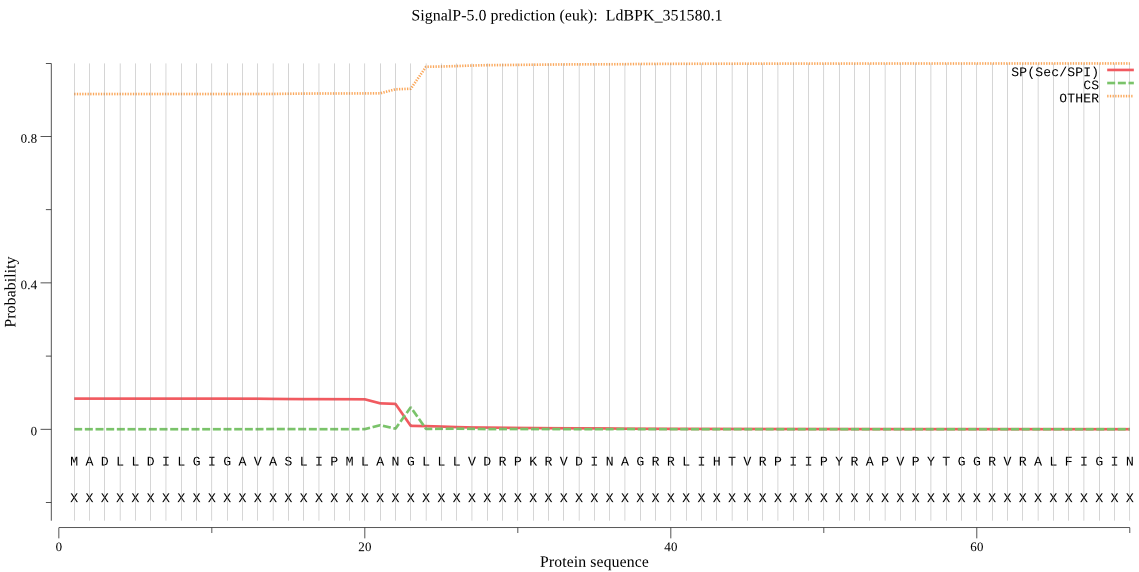
| LdBPK_351580.1 | OTHER | 0.583777 | 0.412160 | 0.004063 |
MADLLDILGIGAVASLIPMLANGLLLVDRPKRVDINAGRRLIHTVRPIIPYRAPVPYTGG RVRALFIGINYTGMRNALRGCVNDVSSMLGTLQQISFPISECCILVDDPSFPGFCGMPTR DNIIKHMLWLTGDVRPGDVLFFHFSGHGGQTKATRDSEEKYDQCLIPLDHVKNGSILDDD LFLMLVAPLPSGVRMTCVFDCCHSASMLDLPFSYVAPRVGGGGAREYMQQVRRGNFSNGD VVMFSGCTDSGTSADVQNGGHANGAATLAFTWSLLNTHGFSYLNILLKTREELRKKGRVQ VPQLTSSKPIDLYKPFSLFGMITVNASMMHCVPQQYQQRPQSLPPQAMPPPAGYPVHVPP PPQGYYPPPQRPGWGLGYPAPGYPAQGIPVQQATLGVSRCPPSQYLPAPPPALYAPPPPG QHGPPQPPPAQYTFSPLPPR
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LdBPK_351580.1 | 157 S | ATRDSEEKY | 0.997 | unsp |