

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_353880.1 | OTHER | 0.999615 | 0.000169 | 0.000216 |
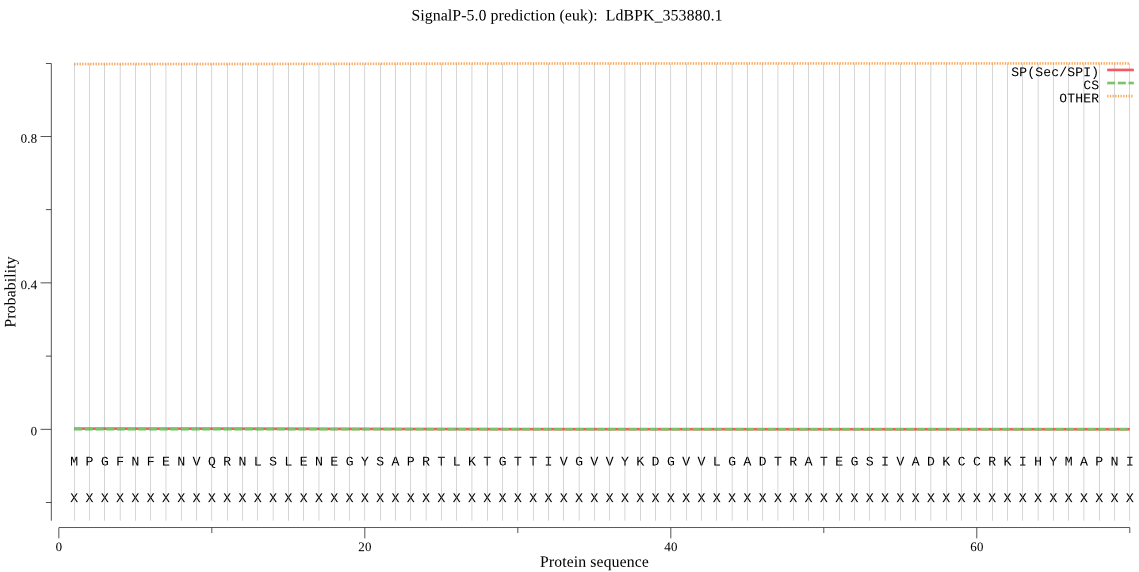
MPGFNFENVQRNLSLENEGYSAPRTLKTGTTIVGVVYKDGVVLGADTRATEGSIVADKCC RKIHYMAPNIMCCGAGTSADTEAVTNMVSSHLALHRLETGKQSRVLEALTLLKRHLYRYQ GHVSAALVLGGVDVEGPFLATIAPHGSTDRLPFVTMGSGSIAAMAQLEVAYKDNMTCEEA KELVTSAIHKGIFNDPYSGTQVDVCVITKDKTELMIGYDTPNKRMHPRQEVMLPPGTTPV LKEEIRHLVDIVDV
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LdBPK_353880.1 | 14 S | QRNLSLENE | 0.991 | unsp |