

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_354910.1 | OTHER | 0.999853 | 0.000087 | 0.000061 |
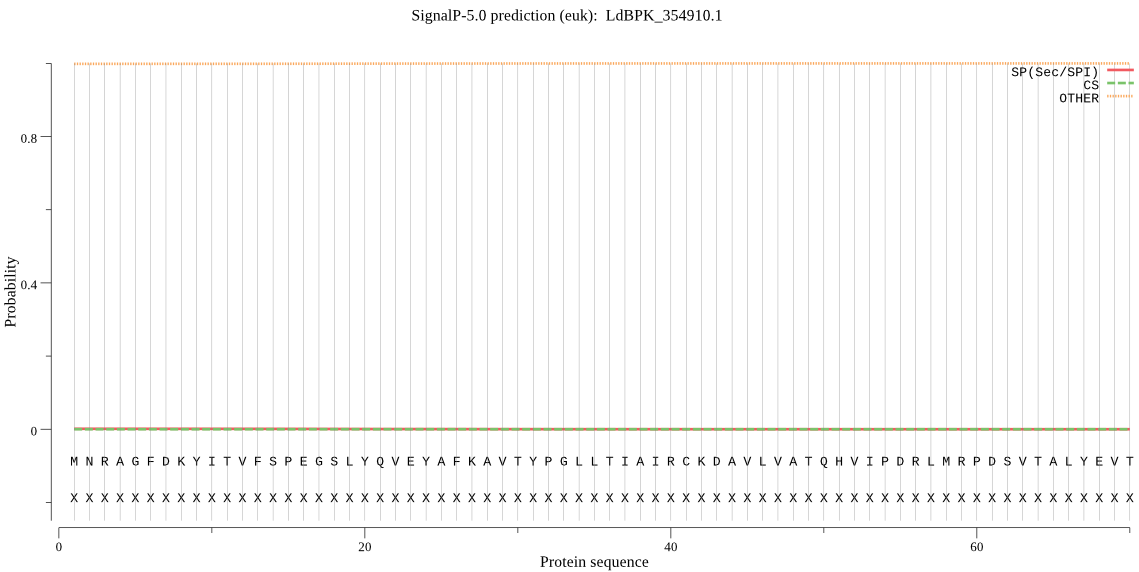
MNRAGFDKYITVFSPEGSLYQVEYAFKAVTYPGLLTIAIRCKDAVLVATQHVIPDRLMRP DSVTALYEVTPSIGCCMTGRAPDGRALVQRAREEASDYHYRYGVQIPIAVLAKRMGDKAQ VRTQQAGLRPMGVVSTFIGMDQSDQDGSLKPQIYTVDPAGWTGGHIACAVGKKQVEAMAF LEKRQKSTKFDELTQKEAAMIALAALQSAIGTAVKAKEVEVGRCTAANPAFQRVPNSEVE EWLTAVAEAD
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LdBPK_354910.1 | 187 S | KRQKSTKFD | 0.998 | unsp |