

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_360220.1 | SP | 0.457809 | 0.522974 | 0.019216 | CS pos: 35-36. VSC-AV. Pr: 0.5207 |
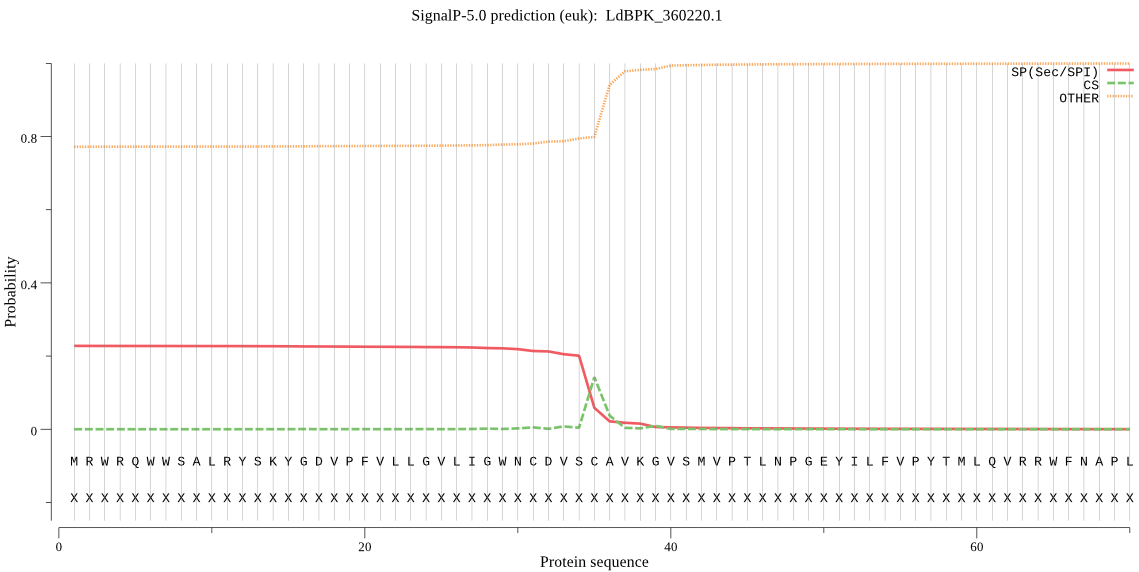
MRWRQWWSALRYSKYGDVPFVLLGVLIGWNCDVSCAVKGVSMVPTLNPGEYILFVPYTML QVRRWFNAPLVNLSDVVVVKVSDDLSVCKRVVKCTSSRAQAEEWGKEHYVEVMPAPYSPP VPQHMNGDDEDSTEVDSATNSERAYFDYVARNTVRSKDWDSCIDRIPNPSQWIWLEGDNK SESFDSRRCGPVPVECIRGLVLASIWPSPHALQRPPPPPRASQLH