

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LdBPK_360340.1 | OTHER | 0.995107 | 0.004806 | 0.000087 |
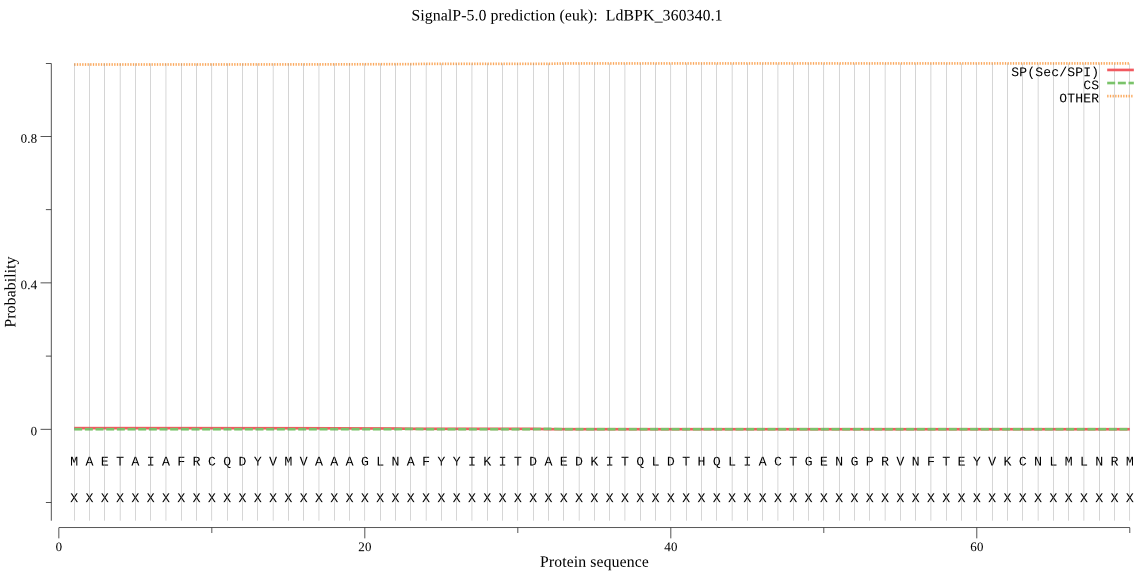
MAETAIAFRCQDYVMVAAAGLNAFYYIKITDAEDKITQLDTHQLIACTGENGPRVNFTEY VKCNLMLNRMRQHGRHSSCDSTANFMRNCLASAIRSREGAYQVNCLFAGYDMPVSEDDDG AVGPQLFYLDYLGTLQAVPYGCHGYGACFVTALLDCLWRPDLTQQEGLELMQKCCDEVKR RVVISNSYFFVKAVTKNGVEVITAVH
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LdBPK_360340.1 | 77 S | HGRHSSCDS | 0.997 | unsp |