

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
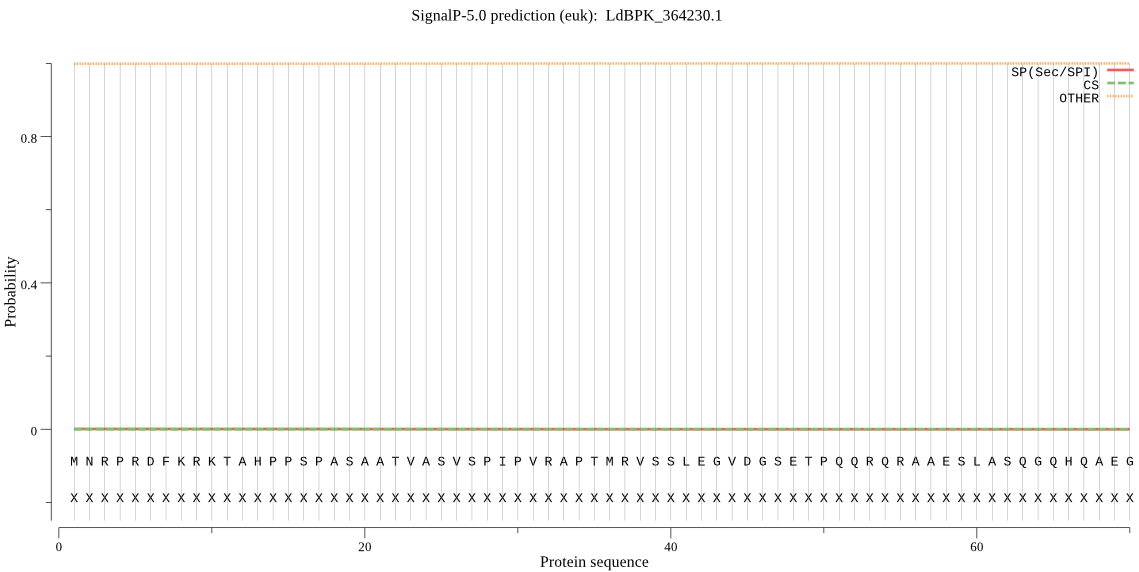
| LdBPK_364230.1 | OTHER | 0.999883 | 0.000031 | 0.000086 |
MNRPRDFKRKTAHPPSPASAATVASVSPIPVRAPTMRVSSLEGVDGSETPQQRQRAAESL ASQGQHQAEGVVKAKSDAQATDIYGSSDSSVPMVAAADACSRASRASPLLSSSEPALSST ARLASVPGIFSSCLASAALRKTKVSEGHSSNQPPTSQQLRQTLRARATVFRPPVRGGAHS PRGGVPKVCPPAPLASGAAAAHGRVASVSTSTPPPASPEALSAHEEHASGVPVSRSSRSR RSSTSALPLSHTYSGAEVQGSNTNVITSSSTNNRGSARSLGNDTVISTAVGRQRPNMAAV LLPTQQLLENGYYLASDESANSSFSDTPRSARRISLGSSSSLRTTTQRLDVNSSGAVGGG RSATVATGASPSGINSLRDSRDPNSYSRGNCSDDNSDIMFEEENDLCNFDEEDEEQAVLT YMDSGAEGNLPGRPSSSNRAPPPPSARQGLLSATGAGDGPSIDPSSLSVSVEVGDTDYDI GEDPPSCMSVSSPPQLHRRGAPALRISACSVRSSAAADWHAPTMVRRPTSVSKASGAPSF QAAVTPAKLTTVTAVPRMLSRQHSTATAATSSRPTPLTTAAVHTPLTATSSSPRFASFSP SLQLSSMNEEELRERECKQLSLYEAFFFNPTRSMWLVDDHVLAKVLEYVRIMGYEHPALK RDRLKGQPSLWSSSSAIASPIAPAKSKKKAAGLRNANSTAPQNMHASSSSAAAEAVVEAL AQPGSANASHTLPTAAGGSADSAAFATKHFCLKAGKPRCARQYVNRSTATAASHLFLTFS EAMRWIAEQEYVIGTVLLCCARFIGDPNADGAAVLGDGVADTAPWHPRRALLQRRKPCIP LRFHPIHFAASLVCSRFPQWGGMSAFTRAWETQIRLVLGSASPSHQRLFKLPDDALLLSS DCEAGNLHRVERAGEPYSFLIWLEPDLGSDKRIWFRFSVTGAKEGRRLRFRLMNAAPHVK LYRQNGMMPVWRDGLSQPNWGPVDSCSFRTTNRDLDGEVSFSINPRNSTETIQIAFCAPY TYADLLCHVCHWHALVKSSGCDMRFEERVLCRSPDGRKLHLLIVTSRVGGAPALAATEDK STSDAAISGGRGHTGVGADGVAAAGAGSSSGWTSPGVASPTMTIPPGKGKGGATKEAVRG PYANFASGKKVVLVSGRVHPGEVTASHGVHGLISFLLSSDVRAIQLREHFIFFIVPMLNP DGVSRGHSRMDQFGNNLNRCYNDPDAETQPTVLALRRVFEHLQHTYRERFIMYLDFHSHA SQSSGFMFGNNLPVSVQHWNLFFPRLVELHVRHVFSFALCRFGRVHMTSKDGASRVLFGS SLIHSYTVELPHFTDRRLYADEYAAMNNGSNVLFEVTWPPLHQTSGQAGSVDVNEDGAQD AENGSEGKWRGVRPAVGARGLYKRSNMLLAGRGRACRSNRESSSAQARDGSPPKKSSGAA PRSQGHANSAGVGEHCSGQSSATQIGGTRLASFLQPISTPSILCQSAEVGQACLLALRDY CSIGARPSPELTMFGGMDGVLRDSKRQVKLDSSRKCKKAQSVATYAGINPIYKQY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LdBPK_364230.1 | 180 S | GGAHSPRGG | 0.997 | unsp | LdBPK_364230.1 | 180 S | GGAHSPRGG | 0.997 | unsp | LdBPK_364230.1 | 180 S | GGAHSPRGG | 0.997 | unsp | LdBPK_364230.1 | 217 S | PPPASPEAL | 0.996 | unsp | LdBPK_364230.1 | 237 S | VSRSSRSRR | 0.995 | unsp | LdBPK_364230.1 | 239 S | RSSRSRRSS | 0.995 | unsp | LdBPK_364230.1 | 242 S | RSRRSSTSA | 0.995 | unsp | LdBPK_364230.1 | 243 S | SRRSSTSAL | 0.998 | unsp | LdBPK_364230.1 | 276 S | NNRGSARSL | 0.993 | unsp | LdBPK_364230.1 | 323 S | SANSSFSDT | 0.995 | unsp | LdBPK_364230.1 | 335 S | ARRISLGSS | 0.995 | unsp | LdBPK_364230.1 | 376 S | SGINSLRDS | 0.995 | unsp | LdBPK_364230.1 | 470 S | SLSVSVEVG | 0.994 | unsp | LdBPK_364230.1 | 530 S | RRPTSVSKA | 0.992 | unsp | LdBPK_364230.1 | 592 S | ATSSSPRFA | 0.995 | unsp | LdBPK_364230.1 | 686 S | APAKSKKKA | 0.993 | unsp | LdBPK_364230.1 | 1008 S | NPRNSTETI | 0.995 | unsp | LdBPK_364230.1 | 1039 S | LVKSSGCDM | 0.992 | unsp | LdBPK_364230.1 | 1081 S | TEDKSTSDA | 0.997 | unsp | LdBPK_364230.1 | 1208 S | SRGHSRMDQ | 0.998 | unsp | LdBPK_364230.1 | 1422 S | SNRESSSAQ | 0.995 | unsp | LdBPK_364230.1 | 1508 S | GARPSPELT | 0.996 | unsp | LdBPK_364230.1 | 1524 S | VLRDSKRQV | 0.993 | unsp | LdBPK_364230.1 | 40 S | MRVSSLEGV | 0.995 | unsp | LdBPK_364230.1 | 107 S | ASRASPLLS | 0.99 | unsp |