

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
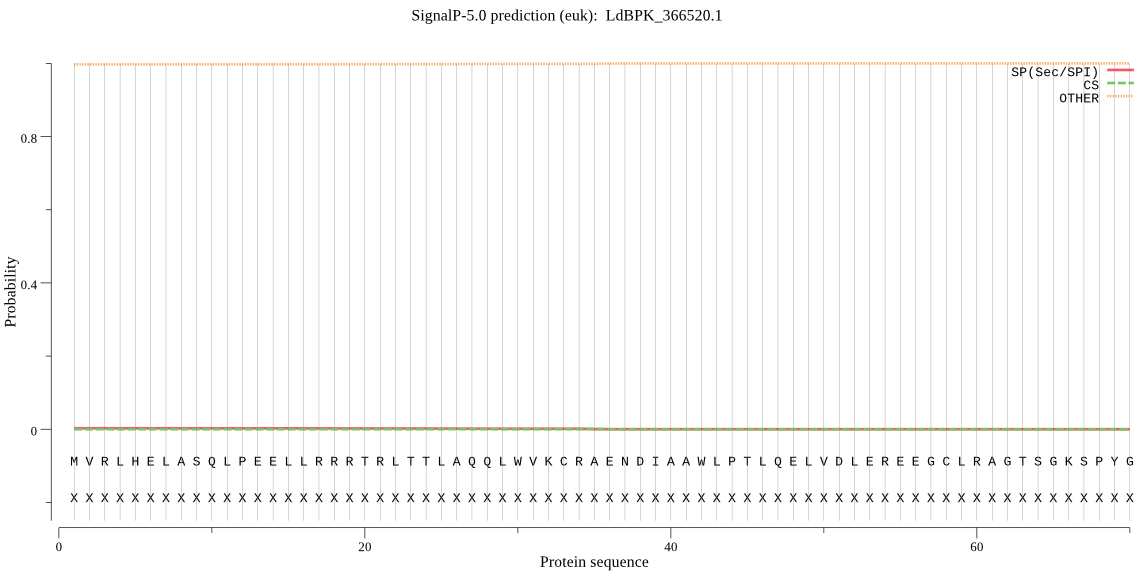
| LdBPK_366520.1 | OTHER | 0.981424 | 0.007667 | 0.010909 |
MVRLHELASQLPEELLRRRTRLTTLAQQLWVKCRAENDIAAWLPTLQELVDLEREEGCLR AGTSGKSPYGALLGANEPGMTVAKLDAIYADIKSWLPALYKEVLENRKDVGASLTELQTP ISKEKQIALGRQLMTDVWKYDWGAGRYDEAPHPFGGMLKEYVRMTYYWSPDNYTKCLLAT IHETGNAKYEQNCGPRELLGQPVCEARSGGIHETQSLLAERMIAMSAAFAEYLAPLLELH LGAQPGLTVKNVRKINQLVKPSCIRKLADEVGYSLSLHVILRYEIERNLIEGRLEAANVP RVWDEKMKEYMGLETLGRDDLGCLKGIHWDAGYWAGFPAYTIGTVAQCLLPPRPQARRAA AEWCSRLSSVL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LdBPK_366520.1 | 67 S | TSGKSPYGA | 0.994 | unsp |