

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmjF.02.0710 | mTP | 0.041427 | 0.005178 | 0.953395 | CS pos: 62-63. CRW-QS. Pr: 0.3121 |
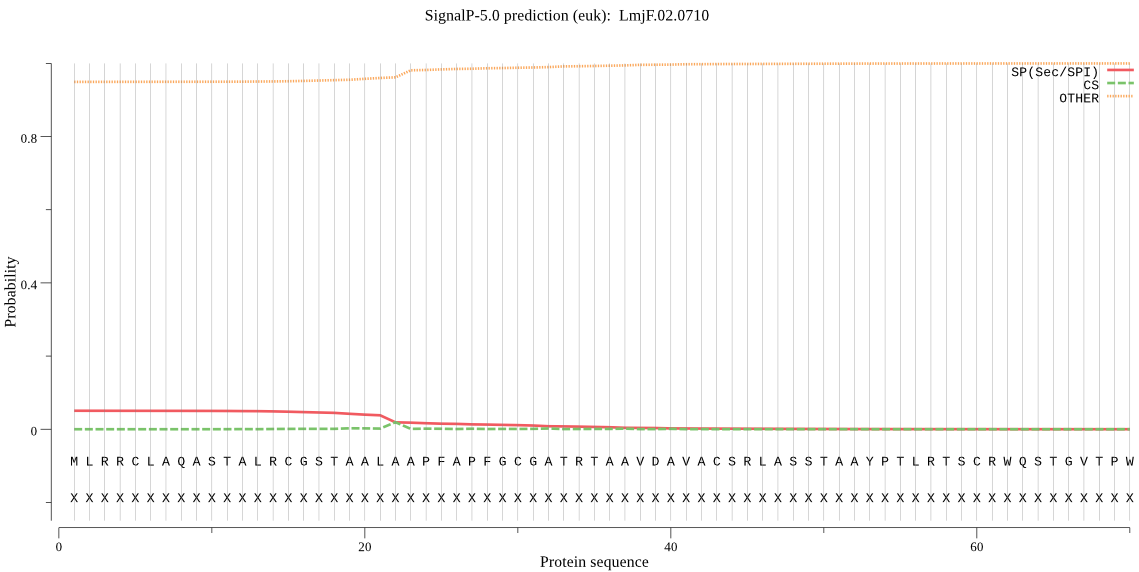
MLRRCLAQASTALRCGSTAALAAPFAPFGCGATRTAAVDAVACSRLASSTAAYPTLRTSC RWQSTGVTPWKSSGNSGGGGNGGQQQGRTWVNPNAVPPGEFLKKYARDLTEEARTGRLDP IVGREEVIRRTIQVVSRRTKNNPVLIGEPGVGKTAIVEGLAQRIVKGEVPESIKDRQVYA LDMGTLVAGAKFRGEFEERLKGVLKDTIESHGKVILFIDELHTLVGAGSSGDGSMDAANL LKPSLARGELHCIGATTLDEYRQHIEKDAALARRFQSVLVTEPTVEETISILRGIKEKYE AHHGCLIKDEALVYAAVNSHRYLSERRLPDKAIDLIDEAASRLRLQQESKPEALDLIERE LIRLKIEAEAVKKDKDEVGKEKLENLYEQISQKQKEYDALEERWKKEKNLFQTIKQRTEE LDVLRHHLEQAMNSGDFAKAGEIQHSQIPNLLKQIEKDKHLASSKNFMVHDSVTSKDIAE VIARATGIPVAQLMTSEREKLIHMDAELKKTIMGQDAAIESITNVVRISRAGLHSHKRPL GSFLFLGPTGVGKTEVCKSLAKFLFDDESFICRIDMSEYMERHSVHRLIGAPPGYVGYEE GGELTESVRRRPYQIVLFDEFEKAHPSVSNILLQVLDEGHLTDSHGRRVDFKNTIIILTS NIGADVIARLPEGKPSISAMPAVMEQVRQRLAPEFINRLDDIIMFNRLSREDIRSIVEIL FAQVQKMLTEHNITLEESTEVYDWLSVNGYSAVYGARPLKRLVQSELLNQLALMLLDGRV REGEHVRLSVQDGHVVVVANHALSPEKGPTDERLLDQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmjF.02.0710 | 324 S | HRYLSERRL | 0.997 | unsp | LmjF.02.0710 | 324 S | HRYLSERRL | 0.997 | unsp | LmjF.02.0710 | 324 S | HRYLSERRL | 0.997 | unsp | LmjF.02.0710 | 535 S | AGLHSHKRP | 0.995 | unsp | LmjF.02.0710 | 584 S | MERHSVHRL | 0.995 | unsp | LmjF.02.0710 | 709 S | FNRLSREDI | 0.998 | unsp | LmjF.02.0710 | 789 S | HVRLSVQDG | 0.997 | unsp | LmjF.02.0710 | 804 S | NHALSPEKG | 0.997 | unsp | LmjF.02.0710 | 59 S | TLRTSCRWQ | 0.994 | unsp | LmjF.02.0710 | 172 S | EVPESIKDR | 0.997 | unsp |