

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
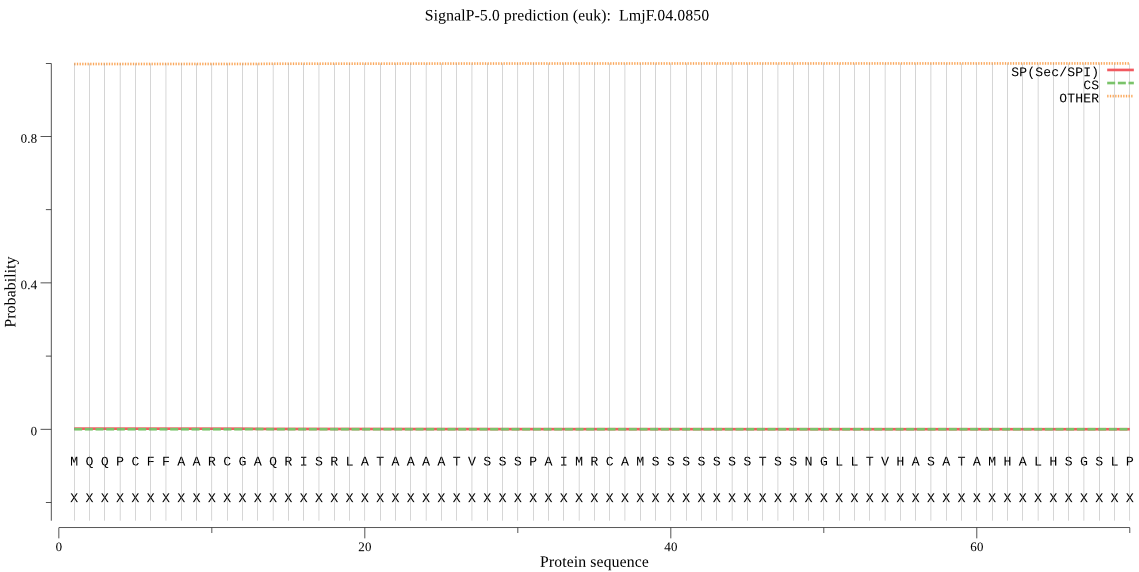
| LmjF.04.0850 | OTHER | 0.730595 | 0.004422 | 0.264983 |
MQQPCFFAARCGAQRISRLATAAAATVSSSPAIMRCAMSSSSSSSTSSNGLLTVHASATA MHALHSGSLPVSSTLTQRNLSALATSLTALRTMRGAATIVVPARASFFAGAAVSPMATAC SSLFAAQRCYGVARATRVALSPKKSNEGSGQLQGVNNLERYKGEEEDEQKNGGGNEQENP FNMNSGPGYLPFPPPFHTINIVMILFLANVMCYLIMNLGNDDWRDFVVEHCTLSHENWTR IYPLFTNAFYQENILQLLIDCWLLWQFGDTMLGFLGNTRMTFFALLCTLGGSVIHVARQK FELYYGMDELEVRGRCYGPNPFIMGLVAIEGLIFRHLNFIQQPPIPFLVLTAFVMVIDVW RIFTTKPEEHGAATGGALMAYLFWALPTRMLGLDKLTATL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LmjF.04.0850 | 141 S | RVALSPKKS | 0.997 | unsp |