

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmjF.08.0450 | OTHER | 0.920334 | 0.076028 | 0.003637 |
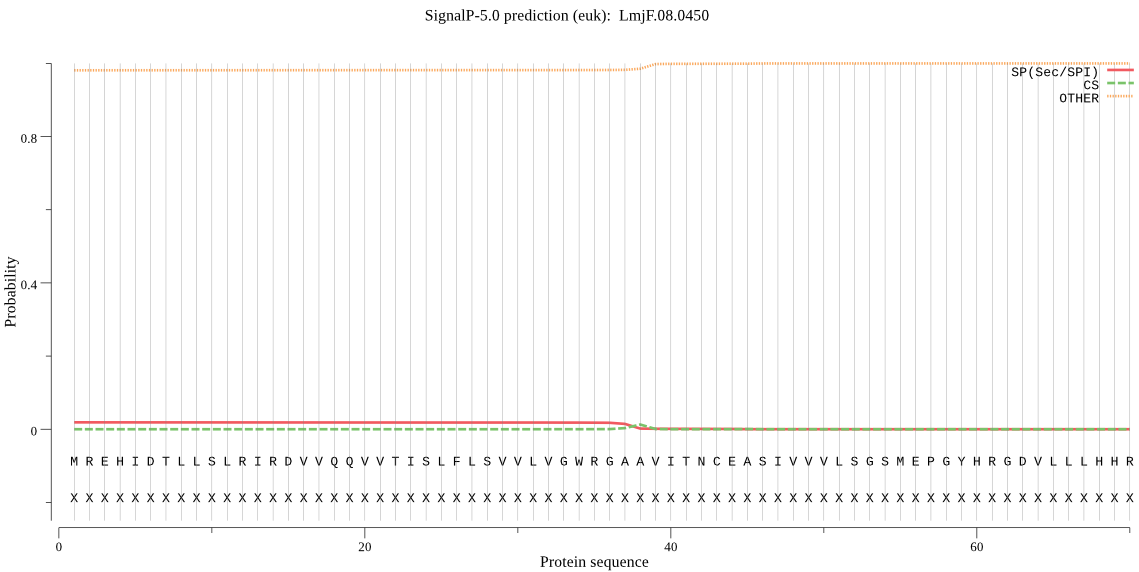
MREHIDTLLSLRIRDVVQQVVTISLFLSVVLVGWRGAAVITNCEASIVVVLSGSMEPGYH RGDVLLLHHRPEYPVEVGDIIVYTLPGQDIPIVHRVHRIHERAEDHKRLYLTKGDNNMND DRFLFHDGREWVEQDMIIGKTFAYVPRIGYLTIVFNESKTIKYVALALLGFFMLTSTQEL