

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
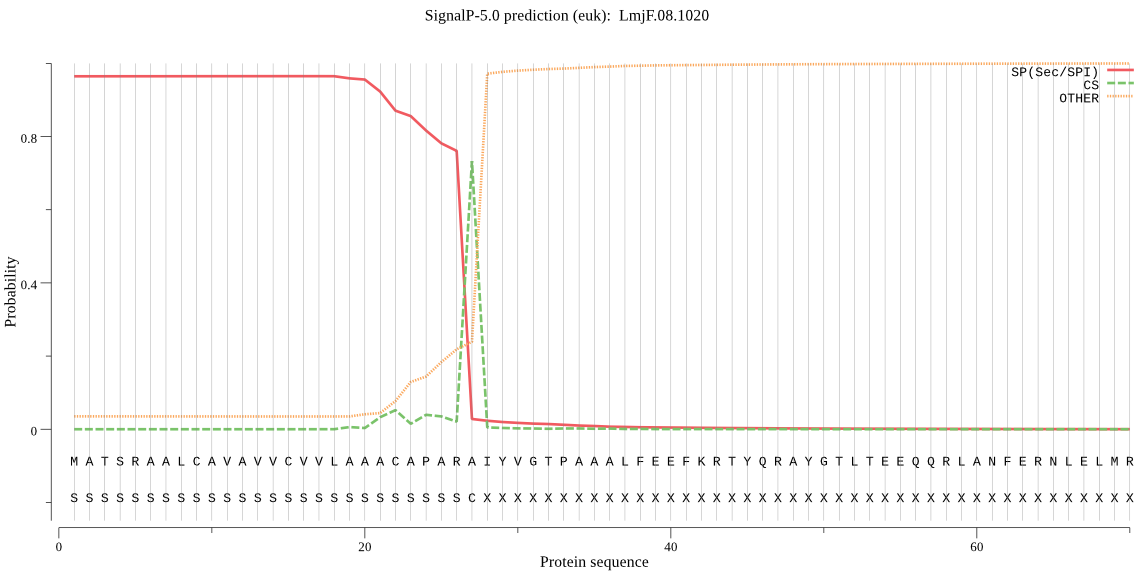
| LmjF.08.1020 | SP | 0.000857 | 0.997237 | 0.001906 | CS pos: 27-28. ARA-IY. Pr: 0.7281 |
MATSRAALCAVAVVCVVLAAACAPARAIYVGTPAAALFEEFKRTYQRAYGTLTEEQQRLA NFERNLELMREHQARNPHARFGITKFFDLSEAEFAARYLNGAAYFAAAKQHAGQHYRKAR ADLSAVPDAVDWREKGAVTPVKNQGACGSCWAFSAVGNIESQWAVAGHKLVRLSEQQLVS CDHVDNGCGGGLMLQAFEWVLRNMNGTVSTEKSYPYVSGNGDVPECSNSSELAPGARIDG YVSMESSERVMTAWLAKNGPISIAVDASSFMSYHSGVLTSCIGEQLNHGVLLVGYNMTGE VPYWVIKNSWGEDWGEKGYVRVTMGVNACLLTGYPVSVHVSQSPTPYL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LmjF.08.1020 | 243 S | DGYVSMESS | 0.995 | unsp |