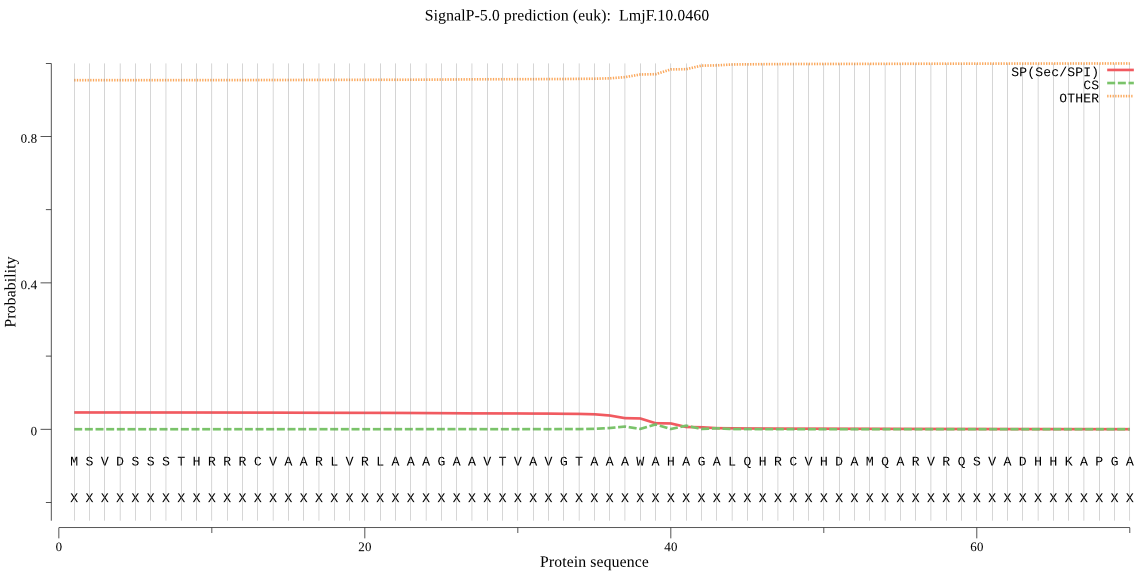
Fasta :-
MSVDSSSTHRRRCVAARLVRLAAAGAAVTVAVGTAAAWAHAGALQHRCVHDAMQARVRQS
VADHHKAPGAVSAVGLPYVTLDAAHTAAAADPRPGSARSVVRDVNWGALRIAVSTEDLTD
PAYHCARVGQHVKDHAGAIVTCTAEDILTNEKRDILVKHLIPQAVQLHTERLKVQQVQGK
WKVTDMVGDICGDFKVPQAHITEGFSNTDFVMYVASVPSEEGVLAWATTCQTFSDGHPAV
GVINIPAANIASRYDQLVTRVVTHEMAHALGFSGPFFKDARIVASVPNVRGKNFDVPVIN
SSTAVAKAREQYGCDTLEYLEVEDQGGAGSAGSHIKMRNAQDELMAPAAAAGYYTALTMA
IFQDLGFYQADFSKAEVMPWGQNAGCAFLTNKCMEQSVTQWPAMFCNESEDAIRCPTSRL
SLGACGVTRHPGLPPYWQYFTDPSLAGLSAFMDYCPVVVPYSDGSCTQRASEAHASLLPF
NVFSDAARCIDGAFRPKATDGIVKSYAGLCANVQCDTATRTYSVQVHGSNDYTNCTPGLR
VELSTVSNAFEGGGYITCPPYVEVCQGNVQAAKDGGNTAAGRRGPRAAATALLVAALLAV
AL