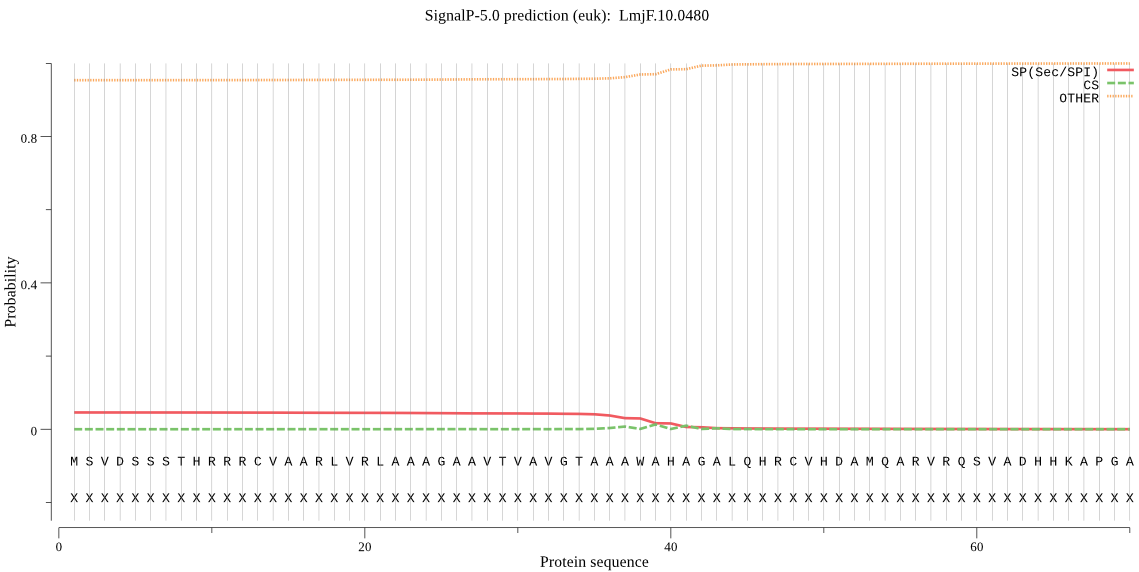
Fasta :-
MSVDSSSTHRRRCVAARLVRLAAAGAAVTVAVGTAAAWAHAGALQHRCVHDAMQARVRQS
VADHHKAPGAVSAVGLPYVTLDAAHTAAAADPRPGSARSVVRDVNWGALRIAVSTEDLTD
PAYHCARVGQHVKDHAGAIVTCTAEDILTNEKRDILVKHLIPQAVQLHTERLKVQQVQGK
WKVTDMVGEICGDFKVPQAHITEGFSNTDFVMYVASVPSEEGVLAWATTCQTFSDGHPAV
GVINIPAANIASRYDQLVTRVVTHEMAHALGFSGPFFEDARIVASVPNVRGKNFDVPVIN
SSTAVAKAREQYGCDTLEYLEMEDQGSAGSAGSHIKMRNAQDELMAAASGAGYYTALTMA
ILQDLGFYQADFSKAEVMPWGQNAGCAFLTNKCMEQNITQWPAMFCNESEDAIRCPTSRL
SLGACGVTRHPGLPPYWQYFTDPSLAGVSAFMDYCPVVVPYSDISCTQRASEAHASLLPF
NVFSDAARCIDGAFRPKATNGIVKSYAGLCANVQCDTATRTYSVQVHGSNDYTNCTPGLR
VELSTVSNAFEGGGYITCPPYVEVCQGNVQAAKDGGNTAAGRRGPRAAATALLVAALLAV
AL