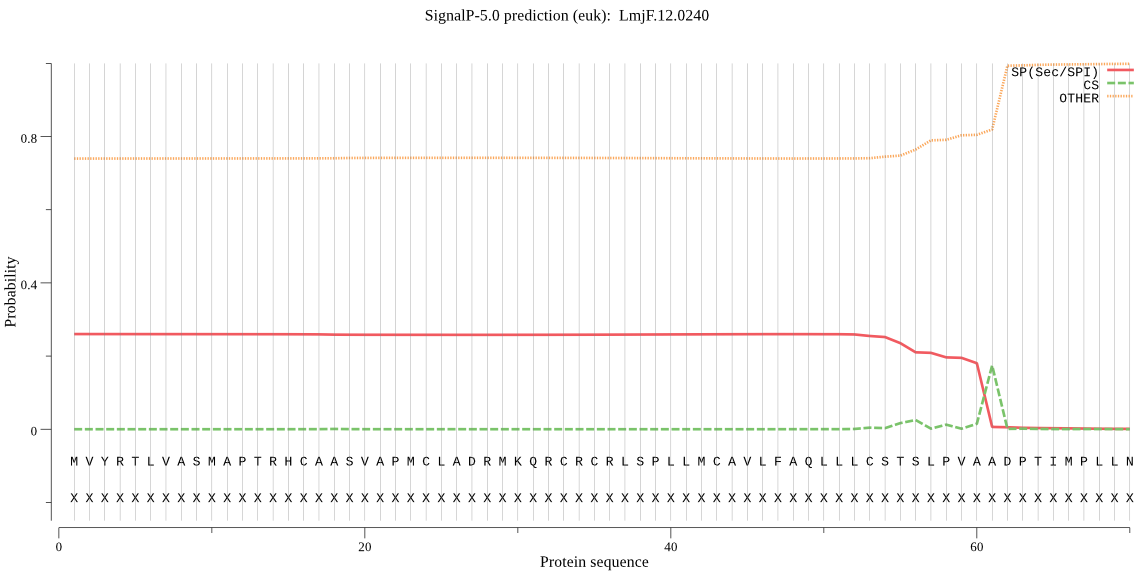
Fasta :-
MVYRTLVASMAPTRHCAASVAPMCLADRMKQRCRCRLSPLLMCAVLFAQLLLCSTSLPVA
ADPTIMPLLNITSTDKADPDEYYKILLSQDATILAGSTTALSSDGVAYPNLNTVKGLRSV
FQVPGAPTVQRAPKRTSTALKAFRADVLSELLVAMALLYLDSTDALPFTRTAPIPSTYLP
ASYTGGASFVNPSFPTVPITLNMLLQHVSSLTKKGLDLRAETAPSGTVPTLSDFVSSLFS
TAATTVFSASQPGLSTSYTYSHANVAIVAYVVEQVLATSTNYSALSGIGEFVFTVVMPPL
QLSNTFLLNRNGHVIDATYPLRSGSSHYTVNYAARQVLDNRSTDILDSAPMDALYFSDYM
LFTTGADVAKLVAEVLVPGGLYHTRLGASMLQNMVPIAVPTMRYTEARTTGLFLFSANKL
CSTMYAAISYSGHAPYCRYSSATVSESAPPFGLVATSGTDELAIVCIPVVSNTKTFCSIA
GLSFSGTGCSRCNAAAAGDRAVGLAMVSLAHLTSEPMPAPTSQSTAHTFNGWFVVLGVAV
TLVVVVVASYITDYLIQPPPPAKIIAPVVASQMGLRSGTALNLPVGGDGAVGSSEVPVDS
ESPETFRKNGRDHDAEDRYSGADNEEQTPHLTRSGGPARGSSSLRRRRPRRRRHRDNGEQ
NPASDGSYSSADDGWGENLDSVDRASTGSQRPNPKGALRFDAYI