

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
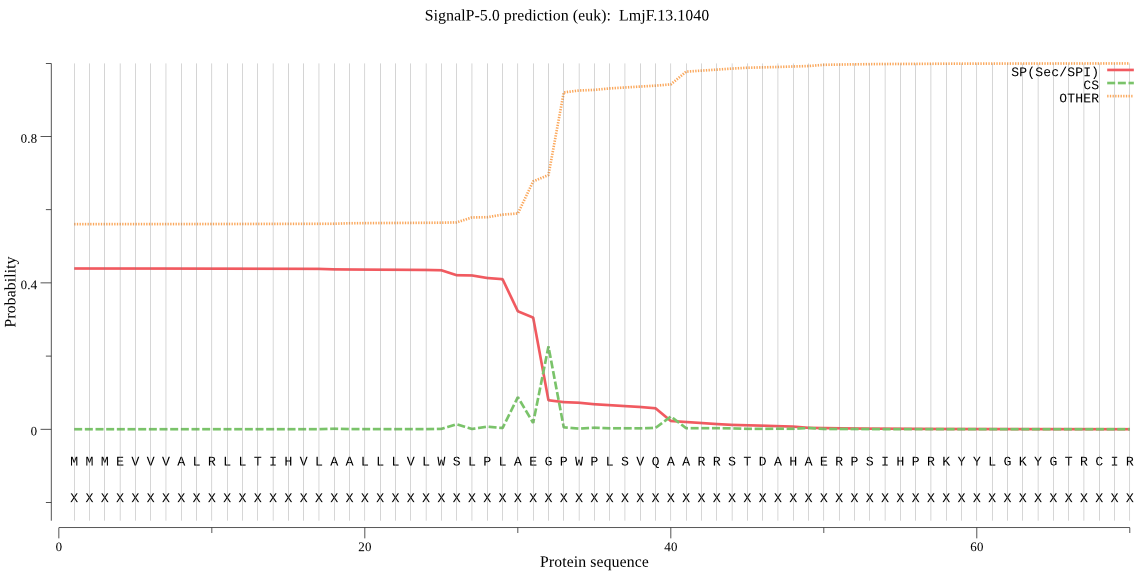
| LmjF.13.1040 | SP | 0.240052 | 0.753693 | 0.006255 | CS pos: 32-33. AEG-PW. Pr: 0.5433 |
MMMEVVVALRLLTIHVLAALLLVLWSLPLAEGPWPLSVQAARRSTDAHAERPSIHPRKYY LGKYGTRCIRNLTGLVVSEDAAERLACFSGRGVRVAVLDTGLCAGITESSRNSVTCTSVV PGVACEDAGCAHGTRSVSVLAGQLSVSTPLPRESLKETAWRNRETHLALADQFIGLAPDS TVRVLRIFDRQGRTNRRHLARALDLLLREAEDGEAGRLHSTWNTSSAHIRRRDETVDVIS LSYGSKDYHSSPQVQDRLYRLMHEHGVIVVAAAGNDGVRFGSVRSPADMPGVLAIGALRV EGHSRSAQTTQASRTLGNLAGSLAAGGGHKSVAHFSGRGPTTWELPFGAGRAKPDLVALG QHVWAVQGVSAAAFSAAAPRARSASSALQLRSASGTSIAAPIVAGVVALCLEAAWSSTSA VASAQNITGGGSVLDVRQNRLSRVSHSLRVREAILRTAVPLEEAAASLPTWPRPAPTTPL HSDSSSTASAAHPRRVLAGVPLLELYAGYLRLSRVSILSQGAGEVQPLRALHAIVAKAAA SSAIDAPEALRSCASFAIPTCVRIGYRIPRCSSNSGDMSTSQSCQPPHDPDGKAHGDSRL PGRPGMSRAAPPAAYWWPFSDQAVYPGATPVLLNISLHLCPPSSSAATDQGARDAAAAAS GEAARRGSTTHDHVTYVITKVSGRLRAREGHPSRNSSHQNDACPLLLFEGAENRHRVGST SPTRLLNLTARSASPPAACTDGEVVGGEATVTGPRHTQAGVCSFAKRRSTQQRWMQHLLR VATELTVVASRSPTRANVVEQLSPPPTSFSLSVAVSSPASAHTHLCYDVARETVAVWRKK GKRSPPSVKGAAASWRSADNNDGHDKQHTKDGAQVPADRDRCVPLFHLFRMLSVEGALHI FTGGSSSQPTLTVPFAVCVVEPPPRVQRVLIDTSLDWFNPTTATSNLFIAGDDPHESDTD GVDAALRRGRQRQRHERAYAEASGGDHPYTNLALLYLYLRHTLGMAVETFPLLHMSTITT SIAANGSSPLWPPAEHTFNASHAAQRAPAGALAHVGTLIIVDPERPLTHGMRRLLTRAVL GGGADRSADGLDVLLVTDWYSADLAAQLHWARDKSLDNAERSSRISGTDAADEVAGVDGD RDAVRELRALRQLENGSTRGLAGSSHVPSWNRWLSEVTVASGSAANSNDCNGAAPLGSEG LLDGKSELPFELSENIVIDGVVVVDAAAPTGRSEPGGNASASHGGTITSAKVVALRSLGQ LNAAGVLRWRLPAQAAAQRQAGRAAAVGASKGVHESNVHSSGHGTCRIGHASNTSAVPST QEGEAVICNVMPGWVQQQRRLVKRTIVSTRDSSAPAAVVEDGWEDVPVAKGDERLVRIGG LEAVDVSTVAPAATDASEAIQSLTHGVLGFLTLPPSSTHVVTASANRFRPGRIAIFTDSD CLSTSNHHAQAALDELEALLYPPSTSPSSPVTTCATWDCLARTPDGQRLLQAESAQSSVC VEVVKELLLWLHTGNLHRWWDSAQLQCEARRWARARHRSTADTTPGADSVGVDRKAANFD EVLGTAPERAHVGEVVGRLWASLVESTERDAASFLVEGPEDETRFQRDYAAHQERTAANV MRALMVNYHGDWRAHLLDHNVSLSSTGAAGDSFSQSLRPASGRRSGIATAQQPEEHVSLI ESLLPLRGMLNALRWLQHPMVLGQLMVSAAVVMSVWLCRAGG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmjF.13.1040 | 285 S | GSVRSPADM | 0.992 | unsp | LmjF.13.1040 | 285 S | GSVRSPADM | 0.992 | unsp | LmjF.13.1040 | 285 S | GSVRSPADM | 0.992 | unsp | LmjF.13.1040 | 442 S | QNRLSRVSH | 0.993 | unsp | LmjF.13.1040 | 542 S | AAASSAIDA | 0.994 | unsp | LmjF.13.1040 | 668 S | ARRGSTTHD | 0.992 | unsp | LmjF.13.1040 | 719 S | HRVGSTSPT | 0.995 | unsp | LmjF.13.1040 | 721 S | VGSTSPTRL | 0.995 | unsp | LmjF.13.1040 | 734 S | ARSASPPAA | 0.995 | unsp | LmjF.13.1040 | 847 S | RSPPSVKGA | 0.997 | unsp | LmjF.13.1040 | 854 S | GAAASWRSA | 0.991 | unsp | LmjF.13.1040 | 957 S | DPHESDTDG | 0.993 | unsp | LmjF.13.1040 | 983 S | YAEASGGDH | 0.992 | unsp | LmjF.13.1040 | 1126 S | SSRISGTDA | 0.998 | unsp | LmjF.13.1040 | 1319 S | SAVPSTQEG | 0.993 | unsp | LmjF.13.1040 | 1348 S | RTIVSTRDS | 0.996 | unsp | LmjF.13.1040 | 1469 S | TSPSSPVTT | 0.994 | unsp | LmjF.13.1040 | 1539 S | ARHRSTADT | 0.997 | unsp | LmjF.13.1040 | 1661 S | LRPASGRRS | 0.997 | unsp | LmjF.13.1040 | 1665 S | SGRRSGIAT | 0.995 | unsp | LmjF.13.1040 | 113 S | SSRNSVTCT | 0.996 | unsp | LmjF.13.1040 | 154 S | LPRESLKET | 0.998 | unsp |