

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmjF.13.1140 | OTHER | 0.981428 | 0.000265 | 0.018307 |
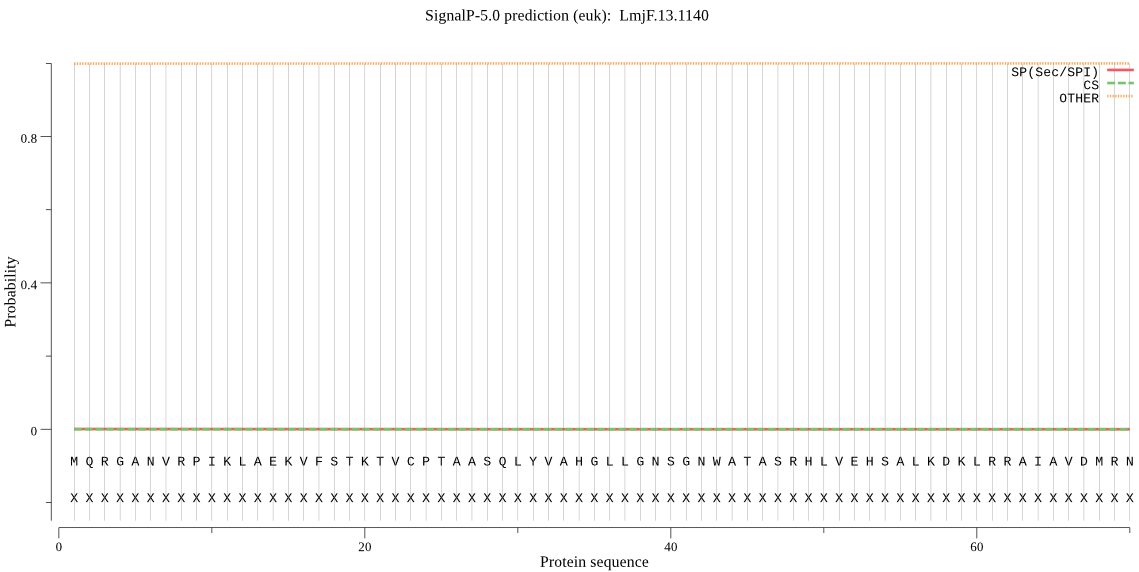
MQRGANVRPIKLAEKVFSTKTVCPTAASQLYVAHGLLGNSGNWATASRHLVEHSALKDKL RRAIAVDMRNHGSSTHSSDHTNAALASDLEALVLREQQELDRAFCDPSSCTTRNAILIGH SMGGLTVMGALLRRANEHHLLTSLVDHCDDVGSTSSYGAWPAEHRRQCIAGMRAVNEEFG FASSQPIAEVLFNNCRASSSLSTGSGNASRIPLLGRVAGAIIVDVTPTMRLGEQRSCADN VKETLERMTQVRLDEIHSYKDATAELIRVGMADEEMRAFVATNIALDPKDKSKPAQWKCN LPVLAGDYGSFQPSITSWFTSSATTMGPASSAAAPRLCTLPVMFVFGGESPYNEPDHRQH ISRFFSNATQVEVAGAGHFVHYEKPKEFTDAVAPFIASLISQP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| LmjF.13.1140 | 74 S | NHGSSTHSS | 0.993 | unsp | LmjF.13.1140 | 258 S | DEIHSYKDA | 0.997 | unsp |