

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
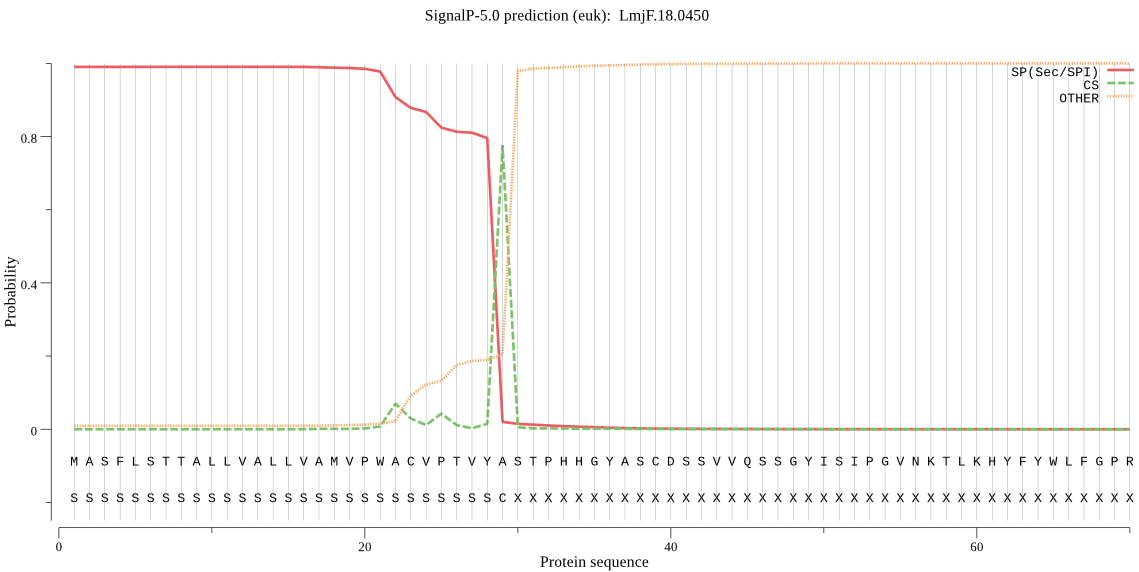
| LmjF.18.0450 | SP | 0.000487 | 0.999502 | 0.000011 | CS pos: 29-30. VYA-ST. Pr: 0.7515 |
MASFLSTTALLVALLVAMVPWACVPTVYASTPHHGYASCDSSVVQSSGYISIPGVNKTLK HYFYWLFGPRKWPKDGREPPVIMWMTGGPGCSSSMALLMELGPCMMNETSGELEHNTYGW NAEAYLLFVDQPTGVGYSYGDTFNYVHNESEVAEDMYNFLQLFAQRFTSPSITGANDFYI IGESYGGHYVPAVSYRILMGNERGDGLRINLKGIAIGNGLTDPYTQLPYHAQTAYYWCKE KLGAPCITEKAYEEMLSLLPACLEKTKKCNEGPDDSDVSCSVATALWAEYVDHYYATGRN SYDIRKQCIGDLCYPMQNTIDFYHKPTVRASLGASAEAQWSTCNNEVSALFERDYMRNFN FTFPPMLDMGIRVLIYAGDMDFICNWLGNEAWVKALRWFGTDRFNAAPNVEFAVSGRWAG LERSYGGLSFVRIYDAGHMVPMDQPEVALFMVHRFLRGQSLA
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LmjF.18.0450 | 138 S | GVGYSYGDT | 0.996 | unsp |