

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmjF.19.0130 | OTHER | 0.999624 | 0.000127 | 0.000249 |
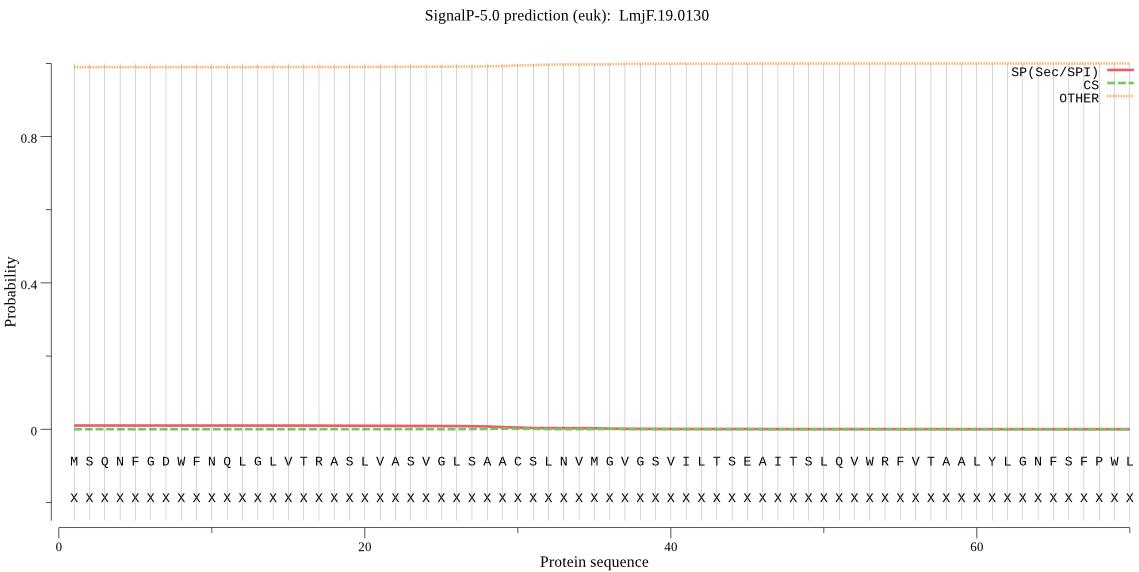
MSQNFGDWFNQLGLVTRASLVASVGLSAACSLNVMGVGSVILTSEAITSLQVWRFVTAAL YLGNFSFPWLMTVAMFVTYVKNNEESDFKGKTADMAYMFLLLLGVLSSAGLFFNVYVTSF SFLMALCWIFCKRHPEQELTLFGFSFRSAVFPWVLMALHLVMGQGLLADILGIVAGHAYV FFKDVLPVSHNQRWLETPMWLRHQFPQPTHRVASFGPEVHPYDPRFQAAWRGEAQQRSSG SHNWGRGQTLGSS