

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmjF.19.1420 | SP | 0.020169 | 0.979020 | 0.000811 | CS pos: 22-23. CYG-SA. Pr: 0.3996 |
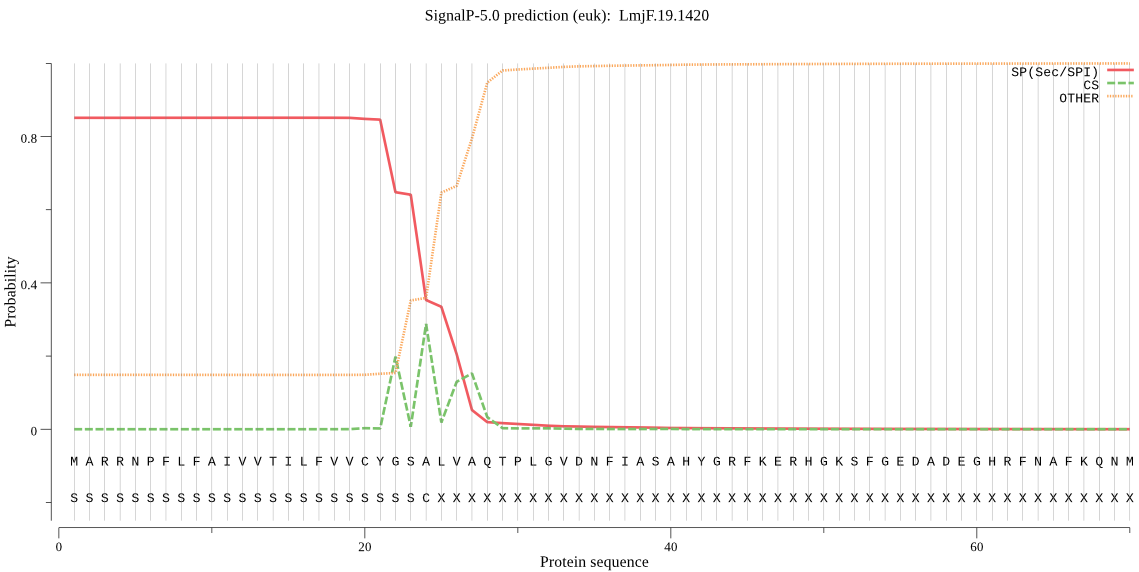
MARRNPFLFAIVVTILFVVCYGSALVAQTPLGVDNFIASAHYGRFKERHGKSFGEDADEG HRFNAFKQNMQTAYFLNTHNPHAHYDVSGKFADLTPQEFAKLYLNPDYYAHRGKDYKEHV HVDDSVLSGAMSVDWREKGAVTPVKNQGMCGSCWAFSAIGNIESQWALKNHSLVSLSEQM LVSCDDIDDGCNGGLMDQAMEWIIQHHNGTVPTEKSYPYASAGGTSPPCHDKGEFGARIS GYMSLPHDEKAIAAYVEKKGPVAVAVDATTWQLYFGGVVTLCFGLSLNHGVLVVGFNKRA KPPYWIVKNSWGTSWGEKGYIRLAMGSNQCLLKNYPVTATVDDSNTSHVPTTTA