

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
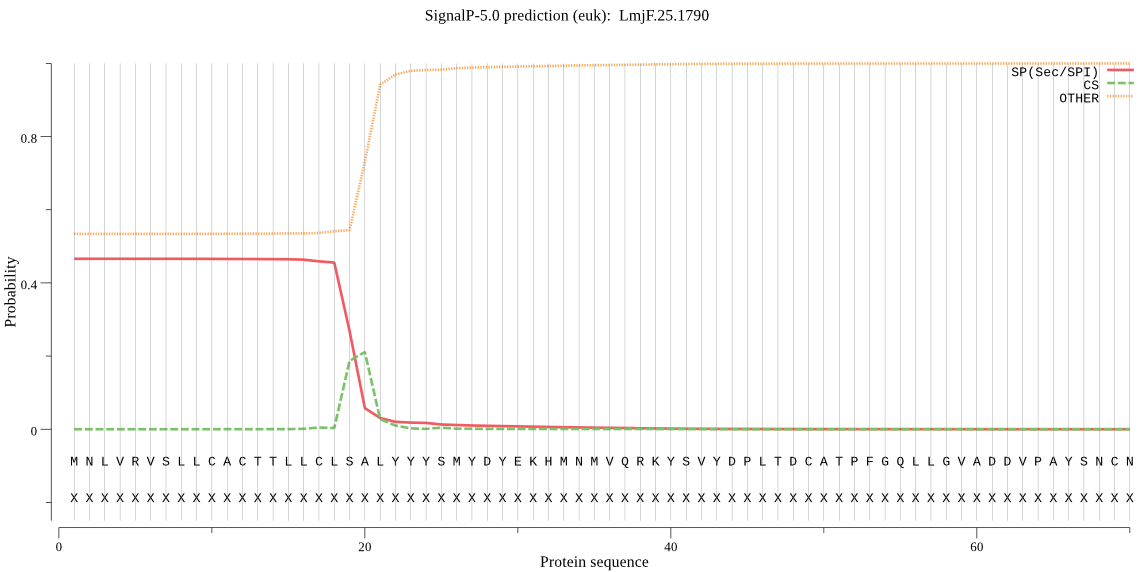
| LmjF.25.1790 | SP | 0.049882 | 0.950014 | 0.000104 | CS pos: 19-20. CLS-AL. Pr: 0.4379 |
MNLVRVSLLCACTTLLCLSALYYYSMYDYEKHMNMVQRKYSVYDPLTDCATPFGQLLGVA DDVPAYSNCNTKFSSTYINYVNLMDPMDNGRRGDPSETRIVMTAYRYTAFDYCMRWLVWN RGVMPRLVENTNQLWKTVDYFNPARPEQGWSAEYITNYEEVTDVEERKFNAPRRGDAIVY RMDKNTIPAGHMAVVVKVEDDVEAAGGPEKLNELKKMRLHPRRVYVAEQNWKNQPWGGHN YSRVLQFKWRAVSEKAHEGGYVDPDELDIIGVVRVGKAMPLRAAPDPYEEALNMDNDGDL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmjF.25.1790 | 261 Y | HEGGYVDPD | 0.991 | unsp | LmjF.25.1790 | 261 Y | HEGGYVDPD | 0.991 | unsp | LmjF.25.1790 | 261 Y | HEGGYVDPD | 0.991 | unsp | LmjF.25.1790 | 41 S | QRKYSVYDP | 0.997 | unsp | LmjF.25.1790 | 253 S | WRAVSEKAH | 0.997 | unsp |