

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmjF.26.2690 | OTHER | 0.822416 | 0.173068 | 0.004516 |
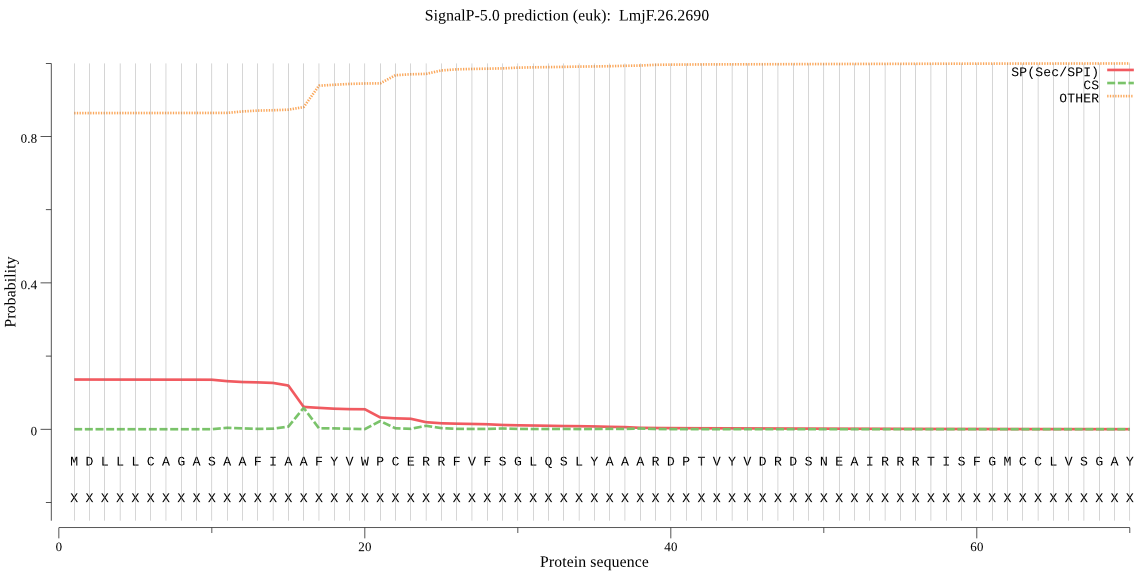
MDLLLCAGASAAFIAAFYVWPCERRFVFSGLQSLYAAARDPTVYVDRDSNEAIRRRTISF GMCCLVSGAYLAFGAAPPRTAHRQSSLLRPIIRSSVSTLLLFAGPIADACHQGTFDTPES TLRLWRNYVICPIGEELFYRGVLFSLLHRRSSPARIFVSAFLFSLSHMHHLVSMACDAYR NGEDKGVAGSDRDEKERACWRSAGHAMCGIVVFTALFGLLSGYYYEHVCERSIIAIAAAH ALCNAIGPPEFMVLRSRHCTVREKVASAAIYVAGIVGWAWTLWRY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmjF.26.2690 | 190 S | GVAGSDRDE | 0.996 | unsp | LmjF.26.2690 | 190 S | GVAGSDRDE | 0.996 | unsp | LmjF.26.2690 | 190 S | GVAGSDRDE | 0.996 | unsp | LmjF.26.2690 | 260 T | SRHCTVREK | 0.991 | unsp | LmjF.26.2690 | 49 S | VDRDSNEAI | 0.996 | unsp | LmjF.26.2690 | 152 S | HRRSSPARI | 0.997 | unsp |