

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
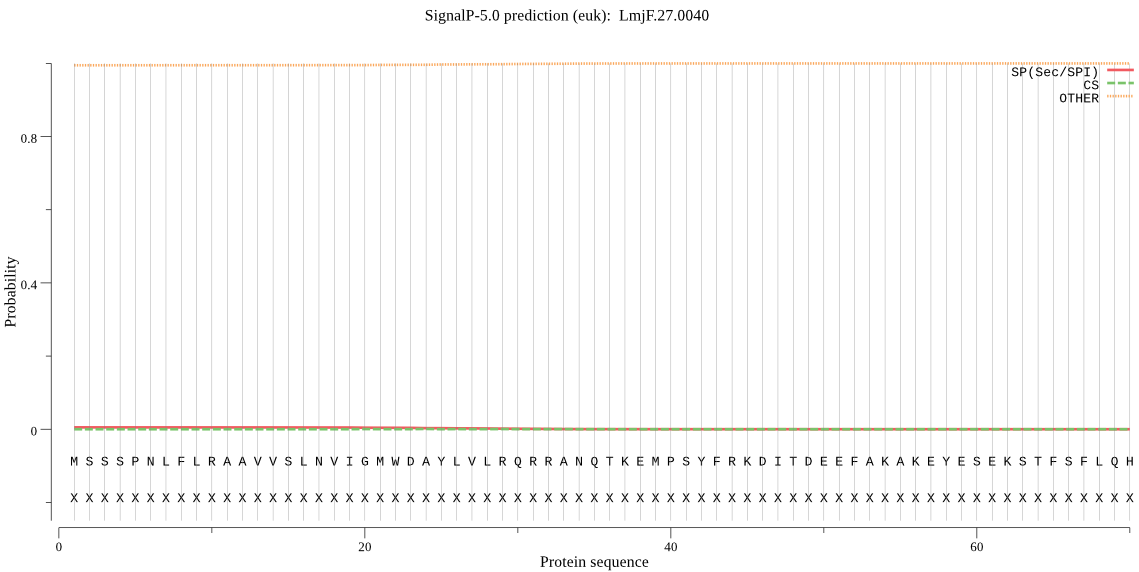
| LmjF.27.0040 | OTHER | 0.987092 | 0.005153 | 0.007756 |
MSSSPNLFLRAAVVSLNVIGMWDAYLVLRQRRANQTKEMPSYFRKDITDEEFAKAKEYES EKSTFSFLQHLKGLVLTNMGIFLRLPALLYYLVAQRASLSTGSFSHNYAAAVAGELISVV LDIPFSYYENFHIEDRHGLNEMTKTEFVKDIVKTLLLRVTLLYPMQIKLIQFVVQRFGER FPLYLFFGMSVMLVVFLLAMPTVIQPLFNKFTPLDAESPLYKKIELLSKEMSFPLKKVFV VDGSRRSHHSNAYFYGFGSNKRIVLYDTILEQLKDDDEPIIAVLCHELGHWKHNHIYVNL AMALGQLMLISYGARLVVFDKRVYEALGFREVDPVVGLNIFAEMFYEPLSTFIGYGFCYV SRRHEFQADRFAVTHHHGEGMKKALLVISKENRASLTPDPLYSALHYTHPPVLERLQAID AELKKRE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| LmjF.27.0040 | 60 S | KEYESEKST | 0.997 | unsp | LmjF.27.0040 | 247 S | GSRRSHHSN | 0.993 | unsp |