

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmjF.28.0570 | SP | 0.001350 | 0.998625 | 0.000026 | CS pos: 24-25. GAA-QG. Pr: 0.4532 |
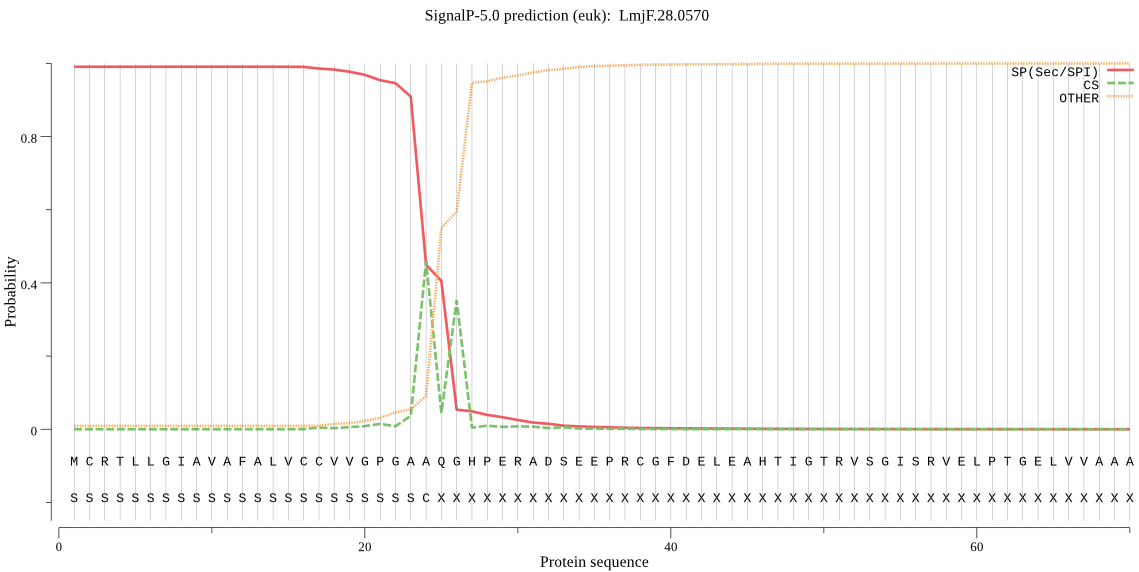
MCRTLLGIAVAFALVCCVVGPGAAQGHPERADSEEPRCGFDELEAHTIGTRVSGISRVEL PTGELVVAAAATGALQPIRIAVFTDDISNSSQHCTASGQSRPNFRGSRVTCSAAEVLTRA KKRVLLELLIPSAVQLHQERLNVQRVNGNIVVDSSIQKDRVCGQFSIREEHMKTGVKDAD FVLYMSAAPTSGSVIAWALKCQNFDNGRPSVGVVNISPKYIAADPKTVRVIAHEVLHALG FSRSVFQERNMLAMASFRSKGPSPVICSEKVVAKAQQHYGCKTQAFMELEDTGDIDDASS HWKRRNAKDELMAGFSGVGIYSALTIAAMEDTGYYQGNYAKAEPMAYGHDAGCKLSSDQC VTNSTSQIPGMFCDAPDAPWSCTSDRLGVGRCILTSHKSNLPTYFQYFSDPRLGGPDPLM DFCPVVEVAEGTMCAATTNALKGSVYGVMSRCVDTPVGFSMDDSAVRQHGICVEVQCDST KYYIKANGASAFCDCPPGSTYNLSTLSPSFSKGYLVCPSYESVCAIKINASLYEEYSGFL TDHSVAGVLTSVKAVVAVLLVVLFMV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmjF.28.0570 | 460 S | PVGFSMDDS | 0.995 | unsp | LmjF.28.0570 | 460 S | PVGFSMDDS | 0.995 | unsp | LmjF.28.0570 | 460 S | PVGFSMDDS | 0.995 | unsp | LmjF.28.0570 | 33 S | ERADSEEPR | 0.996 | unsp | LmjF.28.0570 | 56 S | VSGISRVEL | 0.99 | unsp |