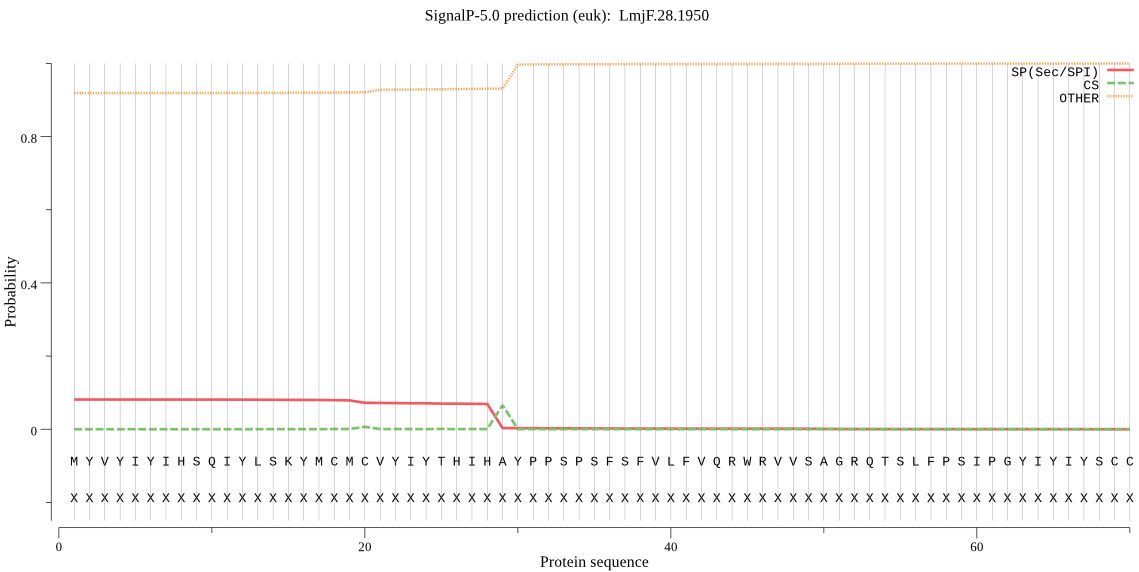
Fasta :-
MYVYIYIHSQIYLSKYMCMCVYIYTHIHAYPPSPSFSFVLFVQRWRVVSAGRQTSLFPSI
PGYIYIYSCCVAIIMRRVCTDLPHPTQFVEHIYITLNDNIRLAARLFMPKDASSEHRYPA
ILEYIPYRKRNGTRIRDEPMHGFFAGHGYVAVRVDMRGAGESDGLLLDEYLKQEQDDALE
VIDWISKQPWCTGDVGMMGKSWGGFNSLQVAARRPPALRAIIVLGFTHNRFTDDIHWKGG
CLLNDNFWWGCIMQGFQSCPPDGDIVGDRWKEMWLERLDKMPLNAADWAEHQRYDAYWQH
GSIQEDYSSIQVPVLLIDGWADSYTNALFHLLEGLKGPCKAICGPWAHVYPQDGTPLPRM
NFLRAAKDWWDYWMKGMTANDVASWPVLQVYIEDSLPPCTIKPMAPGKWVAMDKWVNAEV
PTVMYGLGAGRFLTEMGKDGATTATGTVMVHTPLNHGLISGEWMGVGTIGESAGDQRIDN
GLAVTYTSAPLTAAMDILGQPIFSVTLTCDRPKGFLFAQMCDIAPDGAATRITYGIKNLV
HCGPEGDRAIALVQPGQAVQVTVKMDFCGYCIPAGHSVSLSLANNYWPMVWCSPGDTTLL
LDATTAVFRVPVLHPSATIVPGPDSIPEVAASTPMTVMAPGRVERSVSYEIVLDTWTCVT
SVVGGVFGEGIPRLDDIDTTLEHSLRRELTLSNRDPLSARYTINQKFKVSRPHCVTDVNI
TSTQHCDADYLYIQSKIKAAHNDEGVFEKSFCRRVRREAI