

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
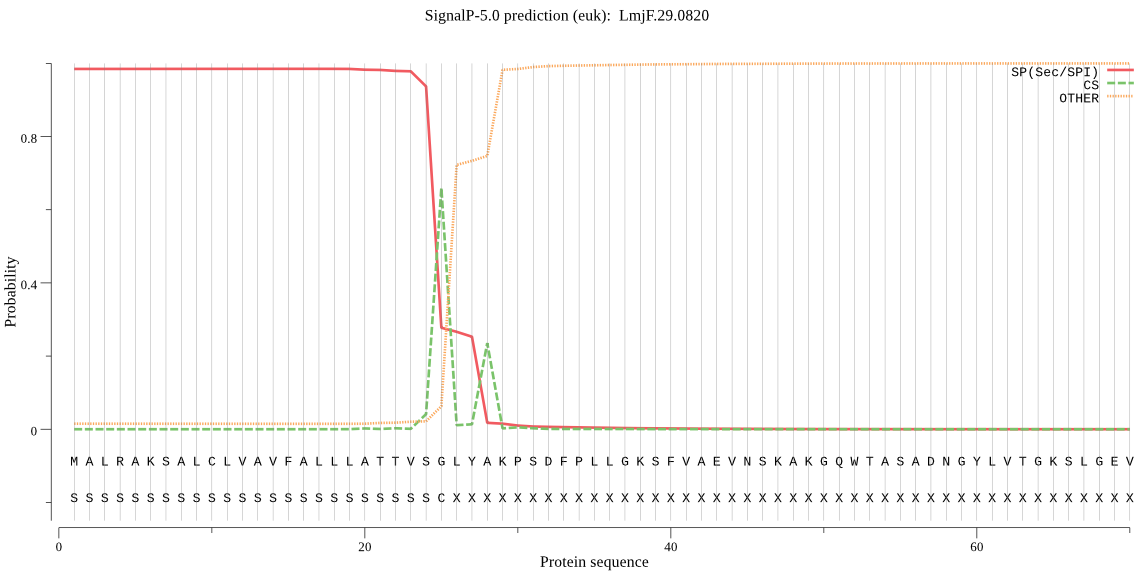
| LmjF.29.0820 | SP | 0.001379 | 0.998582 | 0.000039 | CS pos: 25-26. VSG-LY. Pr: 0.6020 |
MALRAKSALCLVAVFALLLATTVSGLYAKPSDFPLLGKSFVAEVNSKAKGQWTASADNGY LVTGKSLGEVRKLMGVTDMSTEAVPPRNFSVEELQQDLPEFFDAAEHWPMCLTISEIRDQ SNCGSCWAIAAVEAISDRYCTFGGVPDRRMSTSNLLSCCFICGLGCHGGIPTVAWLWWVW VGIATEDCQPYPFDPCSHHGNSEKYPPCPSTIYDTPKCNTTCERSEMDLVKYKGSTSYSV KGEKELMIELMTNGPLELTMQVYSDFVGYKSGVYKHVLGEFLGGHAVKLVGWGTQDGVPY WKVANSWNTDWGDKGYFLIQRGNNECKIESGGVAGIPAQE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmjF.29.0820 | 225 S | TCERSEMDL | 0.993 | unsp | LmjF.29.0820 | 225 S | TCERSEMDL | 0.993 | unsp | LmjF.29.0820 | 225 S | TCERSEMDL | 0.993 | unsp | LmjF.29.0820 | 239 S | STSYSVKGE | 0.993 | unsp | LmjF.29.0820 | 90 S | PRNFSVEEL | 0.996 | unsp | LmjF.29.0820 | 151 S | DRRMSTSNL | 0.997 | unsp |