

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
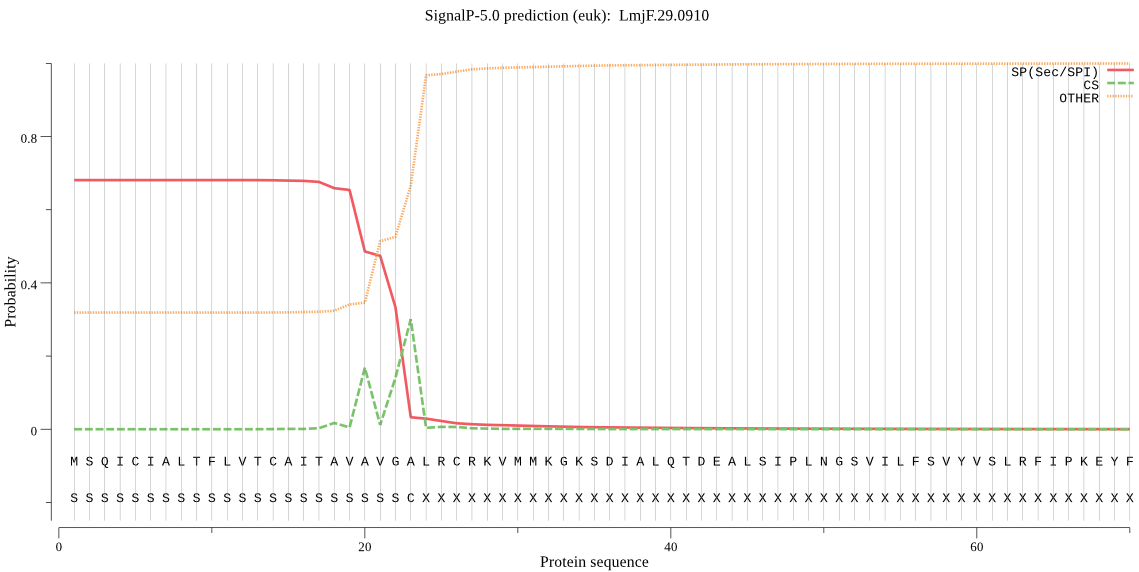
| LmjF.29.0910 | OTHER | 0.921548 | 0.076741 | 0.001712 |
MSQICIALTFLVTCAITAVAVGALRCRKVMMKGKSDIALQTDEALSIPLNGSVILFSVYV SLRFIPKEYFNILISFYLSLISVFALHMFVKGYIKPNIVTGLICVGTGCASFFAQNWIAS NILAFSIAVTALERLPVNGFTTSFILLIGLFFYDIFWVFGSDVMLIVATSIDGPIKLVFP QTIFGDCSKKSLLGLGDIIVPGLFICQTLVFSKDYVKRGSLYFVTSMVAYTLSLVNTMAV MLIFQHGQPALLFIVPWLLVTFSAVAVYNGDVKAAWSFDILSVFTISSEESAPGDPHAEQ EVSLSTFVCEAALDLFGFAKEDSAAESKTAESKEKKEA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| LmjF.29.0910 | 287 S | VFTISSEES | 0.992 | unsp | LmjF.29.0910 | 332 S | KTAESKEKK | 0.995 | unsp |