

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
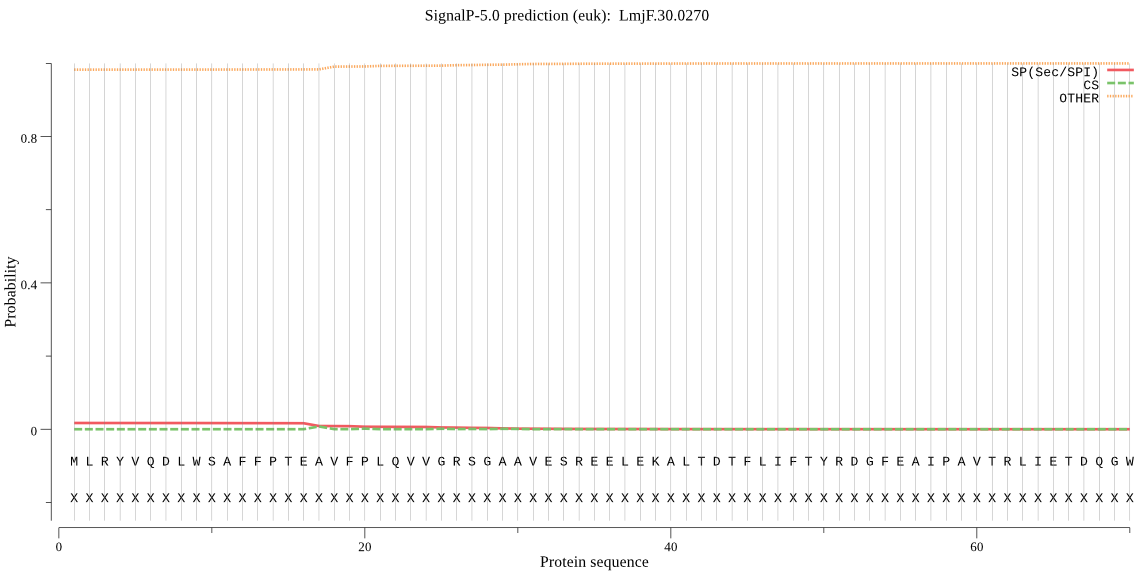
| LmjF.30.0270 | OTHER | 0.952922 | 0.017464 | 0.029614 |
MLRYVQDLWSAFFPTEAVFPLQVVGRSGAAVESREELEKALTDTFLIFTYRDGFEAIPAV TRLIETDQGWGCLLRTSQMLLAHFLWVHGRPADRKLSLFFDHSAETAPFSIHNMIRSVWN RRVFKAEYWSPSQGCEAIKRTVQGAVKTEQLQTRVMVVTSTNGCIYADEVQHTFKQGADV VLVLASVRVSAAAQLTQESYLQIEKLMEQPQCLGVVGGVPGRSYYFFAHNQTQLFYLDPH QRTAAALLCEGLSAAASVTPSVADVRCVHWSRVDTSLFLAFAVTTRDEWAALEVHLSNRF MHVESQQTQRDCDHLGGARPGHADSTFVSPLQMGHSCAMGGASISETAPLVHRRTLRLPR KRFKEGGVGTATAAAAERDDGEDELDTDSWEYLD
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LmjF.30.0270 | 33 S | AAVESREEL | 0.995 | unsp |