

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmjF.32.3680 | OTHER | 0.873720 | 0.000972 | 0.125308 |
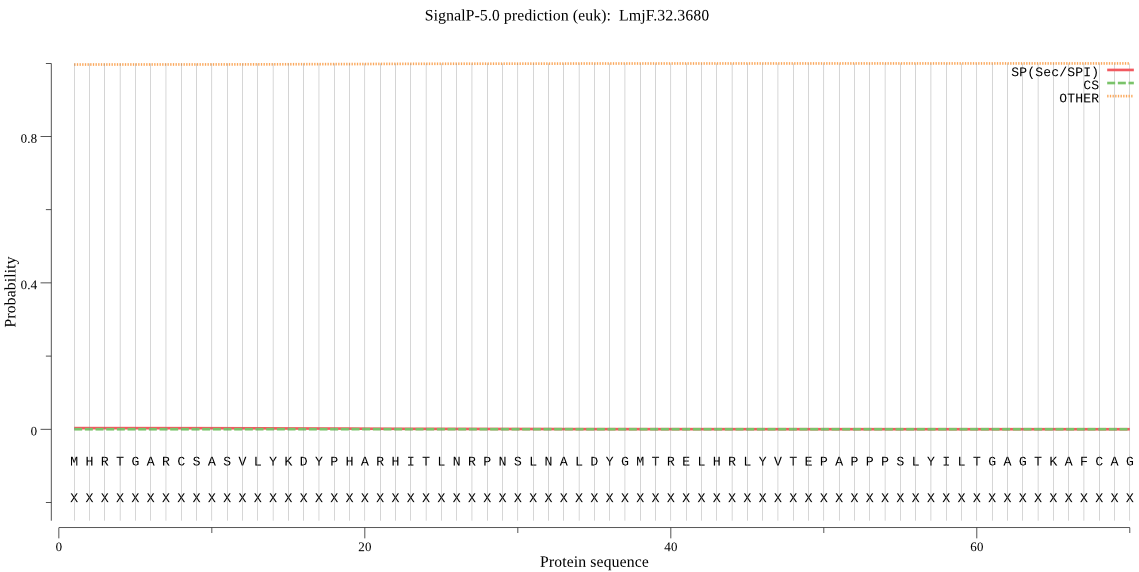
MHRTGARCSASVLYKDYPHARHITLNRPNSLNALDYGMTRELHRLYVTEPAPPPSLYILT GAGTKAFCAGGDVIGLTTNNPPGCGREFFYWEYQVDYKASIIPAGQVCLWDGYVLGGGAG LSIGSAYRVASEKACFAMPEVAIGMFPDVGASWFLPRLSVPGLGLYMGLTGHRLRGADLV HLGLATHFVPSAKMGELEQALVSMSDAGDVEAVLDKYTTPTTQLPPCTIAHSISSLASHF DITADLTISSILDACREHAQTDPLAKAAADLMPSFSPTAMTLALELLKRGAKLSTPVEAF QMEYCVSQRIIAEHDFREGVRALLIDKDKKPKWQPSTVAEVSAEAIDAYFRPTTPDQPVW DPVAPLSERAS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmjF.32.3680 | 203 S | QALVSMSDA | 0.991 | unsp | LmjF.32.3680 | 203 S | QALVSMSDA | 0.991 | unsp | LmjF.32.3680 | 203 S | QALVSMSDA | 0.991 | unsp | LmjF.32.3680 | 131 S | YRVASEKAC | 0.991 | unsp | LmjF.32.3680 | 191 S | HFVPSAKMG | 0.992 | unsp |