

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmjF.33.0400 | OTHER | 0.941523 | 0.000315 | 0.058162 |
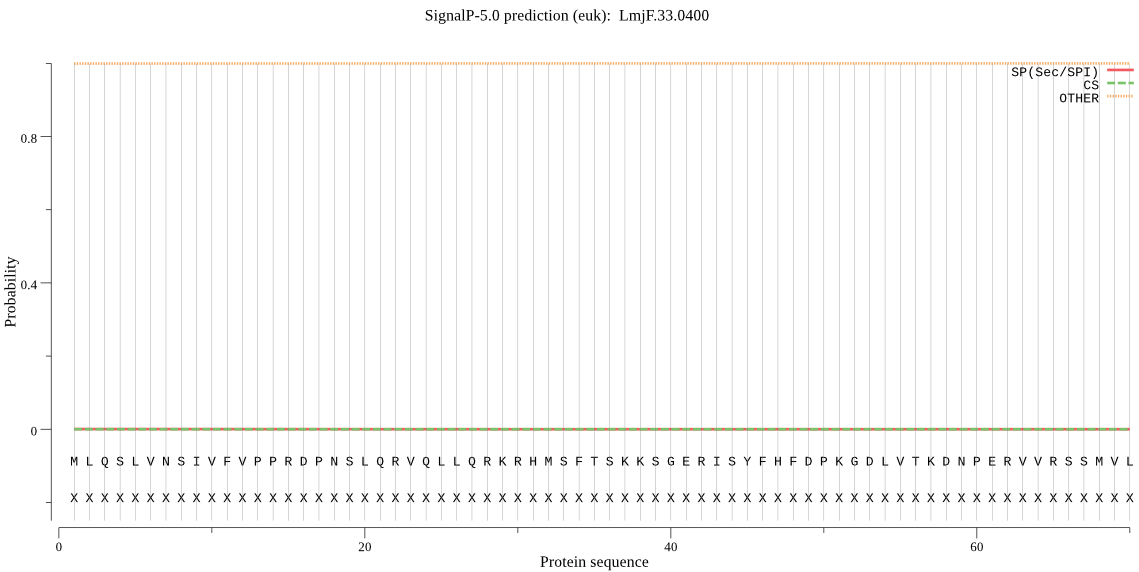
MLQSLVNSIVFVPPRDPNSLQRVQLLQRKRHMSFTSKKSGERISYFHFDPKGDLVTKDNP ERVVRSSMVLLFHHGNAEDLGSAFSYAQSMACVFGVAVVVYDYCGYGFSGFPDAAKPAEV TEKSVYSDADHMYAHLLSLGYPAHRIIIVGRSVGGGPACYLAEKHHKKVGGLVLISTFTS CLRVVSSCCLPYLCCCVDLFPNYRRIEHIMECPVLVMHGTHDNVVPHHCSSELLEDIVAR RNNALQRLLKKREGARANQAKRSSVLGAASTGPTNTTTASATIIVADEALVTAQPPLDGE TVSVFDLYRRAYDGLPEAVRRVAEGRLDVTAADASIGVFHRWFAGCGHNDIEAREGHAFS DMFDYFVRFSTAFSMEREALLLAKPVTGAAKASRDSLNRADSEVRE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmjF.33.0400 | 396 S | ASRDSLNRA | 0.996 | unsp | LmjF.33.0400 | 396 S | ASRDSLNRA | 0.996 | unsp | LmjF.33.0400 | 396 S | ASRDSLNRA | 0.996 | unsp | LmjF.33.0400 | 402 S | NRADSEVRE | 0.992 | unsp | LmjF.33.0400 | 39 S | TSKKSGERI | 0.996 | unsp | LmjF.33.0400 | 374 S | STAFSMERE | 0.991 | unsp |