

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmjF.33.2830 | OTHER | 0.997760 | 0.001550 | 0.000690 |
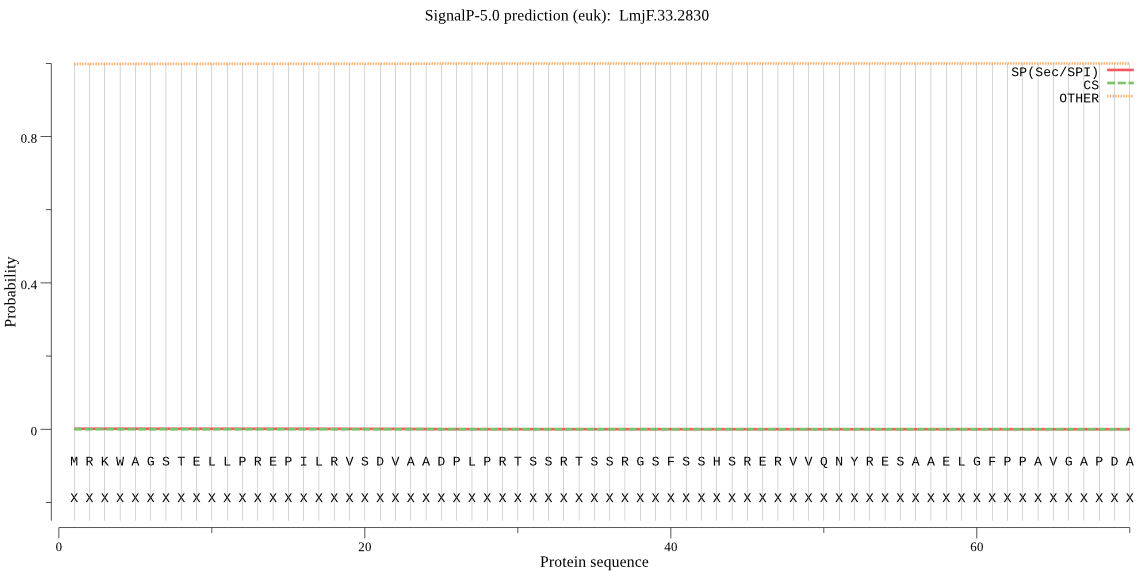
MRKWAGSTELLPREPILRVSDVAADPLPRTSSRTSSRGSFSSHSRERVVQNYRESAAELG FPPAVGAPDAACDDVVVSQSRSHGDTNVQPNDSSKTCGESTRVDDLVIACADPGIASSGA ARCFVLTHDPLLYGILKWSLAVLLLLFLSFIVFGMLYSSIRYGRNCTGTNETVKPLGPCV TPLGTILGVFNGVFGYSNCNDSYVSTELRYINLTVPELNNETGQLTYISKQFYTGLAWQC VEYARRYWMQRGTPQPAYFDSVLGAADVWNLTFVRLLSNASITLPLRRYWNGDRVTDNHQ IPAIGDIIIYPVQDGGFPYGHVAVIANVELSTHGAIYVAEQNWANAVWSSPHHNYTRRIP MFYDMLTSTITLDDSEHQIIGWMRYG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmjF.33.2830 | 39 S | SSRGSFSSH | 0.996 | unsp | LmjF.33.2830 | 39 S | SSRGSFSSH | 0.996 | unsp | LmjF.33.2830 | 39 S | SSRGSFSSH | 0.996 | unsp | LmjF.33.2830 | 44 S | FSSHSRERV | 0.993 | unsp | LmjF.33.2830 | 82 S | SQSRSHGDT | 0.99 | unsp | LmjF.33.2830 | 31 S | LPRTSSRTS | 0.99 | unsp | LmjF.33.2830 | 35 S | SSRTSSRGS | 0.998 | unsp |