

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
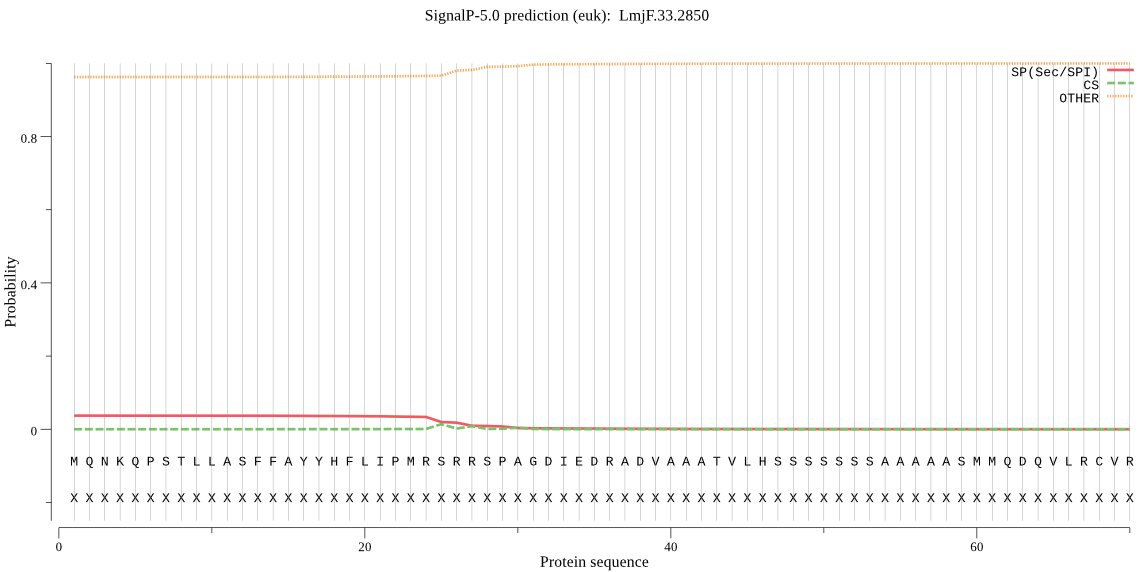
| LmjF.33.2850 | OTHER | 0.879461 | 0.118405 | 0.002134 |
MQNKQPSTLLASFFAYYHFLIPMRSRRSPAGDIEDRADVAAATVLHSSSSSSSAAAAASM MQDQVLRCVRKEPGPSNAAICDAYAMRLLQEATEEERCRGPHWRARCSPSILVALLIWAT IGLVVYMKVDSSHLRPGAALATTSVETPLSQLPESCRGHYCLQEGGKEPFGAVLGAHNGV FAYSNCNSDTCISSLQYQMAIPLPPGARTALDAPHATTRLMTTGMKWQCVEYARRYWMLR GTPTPAFFGAVKGAADIWDSLTHVTFLDNATTAPLLKFKNGARLGYGGSAPRVGDLLIYP RDTENVFPYGHVAVVVKVEMPTKAEADDSYMDAGAASPKPRQRHSHVYVAEQNWDSVTWP NPYHNYSRSLPLVVLESAEGRPLQYTIEDSLHGIQGWVRYDDEP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| LmjF.33.2850 | 28 S | RSRRSPAGD | 0.996 | unsp | LmjF.33.2850 | 329 S | EADDSYMDA | 0.994 | unsp |