

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
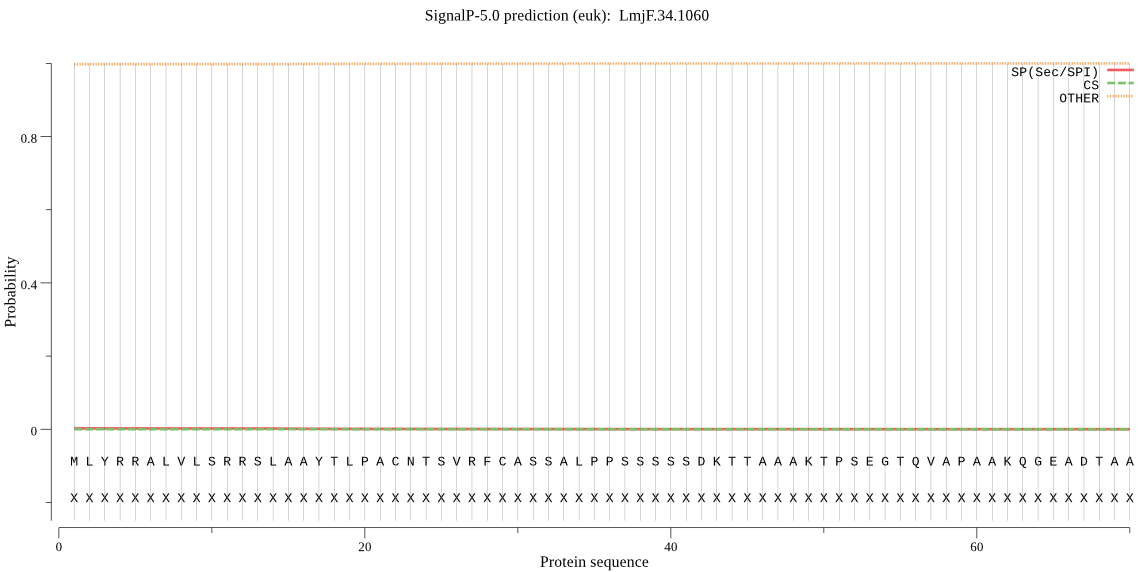
| LmjF.34.1060 | mTP | 0.096880 | 0.000254 | 0.902866 | CS pos: 28-29. VRF-CA. Pr: 0.4675 |
MLYRRALVLSRRSLAAYTLPACNTSVRFCASSALPPSSSSSDKTTAAAKTPSEGTQVAPA AKQGEADTAAADTVAGAGAPSRRGATSPSSDEEGVSAPAPASVSPSSTGAEASTKGLPQL HERGGTSPPEKSTPPAPGANVNDAFAKDRVVPRGFEAFYAKNIVSKMLPKRSPTDAVQRA YTMSPQERVLIAQIAARARLQRTWRHLGILSVLLSALIGYRWYTAQARLDADVSNYKAVV VDVPGHTAVYVDDNGKFIGVRNFIDFATFEKTCDPEKDVLIQFSSYYPWIPMLLLCLLPV LAVVNAGFNGSARMITMAAQAEKSRFTFKREMSVSTRLKDVAGLTEAKHEVVEVIDFLKH PERYQTLGAKLPKGVLLDGPPGVGKTLLAKAVAGEAMVPFVSCSGSEFEEVYVGVGAQRV RELFREAHNCKPCVVFIDEIDAFGRKRRSDGNGSSRGTLNAFLAALDGFKDASGIMVLAA TNRADILDNALTRSGRFDRKISLEKPSYKDRVAIALVHLQPLHLDPSSSLQNYAEMVAAL TPGCSGADIFNVCNEAAIQAAREDKEYVGAPHFHLAVERVLVGLEKSAVKYTPHEKERLA YHEAGIVVLNWFQSDTDPVIKSTILPRGRHRTGVTQKLPQNIYISTQERLLQRIVGRLGG YVSEEHFFSDVSTSAAEDLQMATSMARDMVCVYGMDPVHIGHMGFELDRDDTLQKPFGPE KENAVDIAVETIVQSCLKRARALMLEHLNHTCIIAGLLLKQETLNAHEMWFALGDRPVMT KEFRTYLES
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LmjF.34.1060 | 90 S | TSPSSDEEG | 0.998 | unsp | LmjF.34.1060 | 90 S | TSPSSDEEG | 0.998 | unsp | LmjF.34.1060 | 90 S | TSPSSDEEG | 0.998 | unsp | LmjF.34.1060 | 172 S | LPKRSPTDA | 0.997 | unsp | LmjF.34.1060 | 449 S | RKRRSDGNG | 0.997 | unsp | LmjF.34.1060 | 502 S | DRKISLEKP | 0.993 | unsp | LmjF.34.1060 | 507 S | LEKPSYKDR | 0.997 | unsp | LmjF.34.1060 | 41 S | SSSSSDKTT | 0.995 | unsp | LmjF.34.1060 | 87 S | RGATSPSSD | 0.997 | unsp |