

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmjF.34.4370 | OTHER | 0.898321 | 0.008018 | 0.093661 |
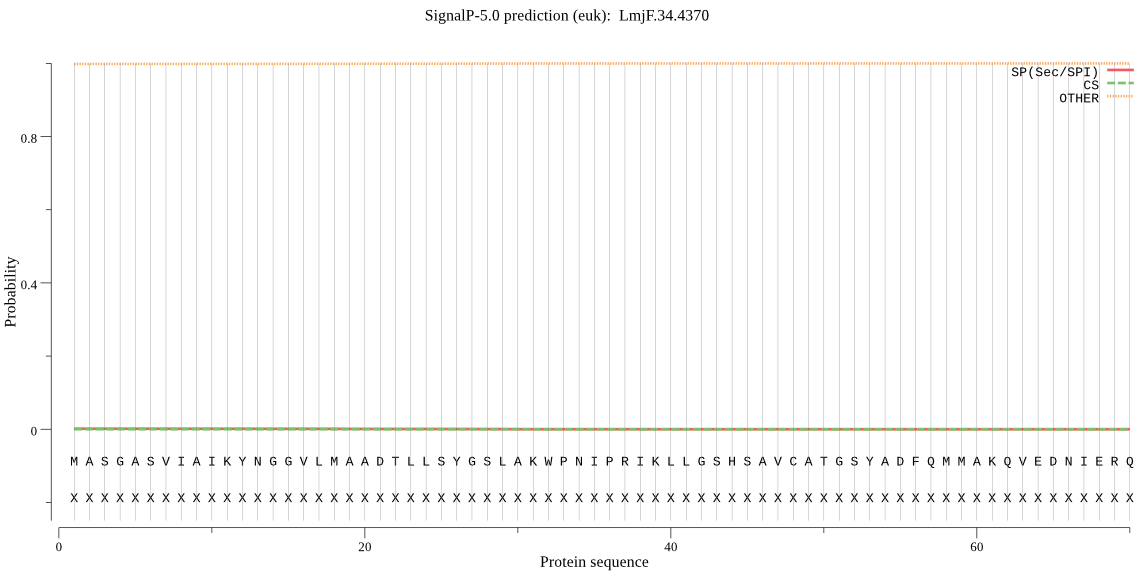
MASGASVIAIKYNGGVLMAADTLLSYGSLAKWPNIPRIKLLGSHSAVCATGSYADFQMMA KQVEDNIERQRMYYNVDELNPSEVFSYLHRSIYQKRCDFEPCLCQMVFIGVRDSETFLAA VDDVGTRWEDDCVATGYGAYIALPLLRQALEKNPGGLSRAQAVQILTDCLRVLFYRECRT INKFQMADAAGDGVRISEPFDVETNWEYEGFCFEKTAIIR