

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmjF.35.1580 | OTHER | 0.616360 | 0.379690 | 0.003950 |
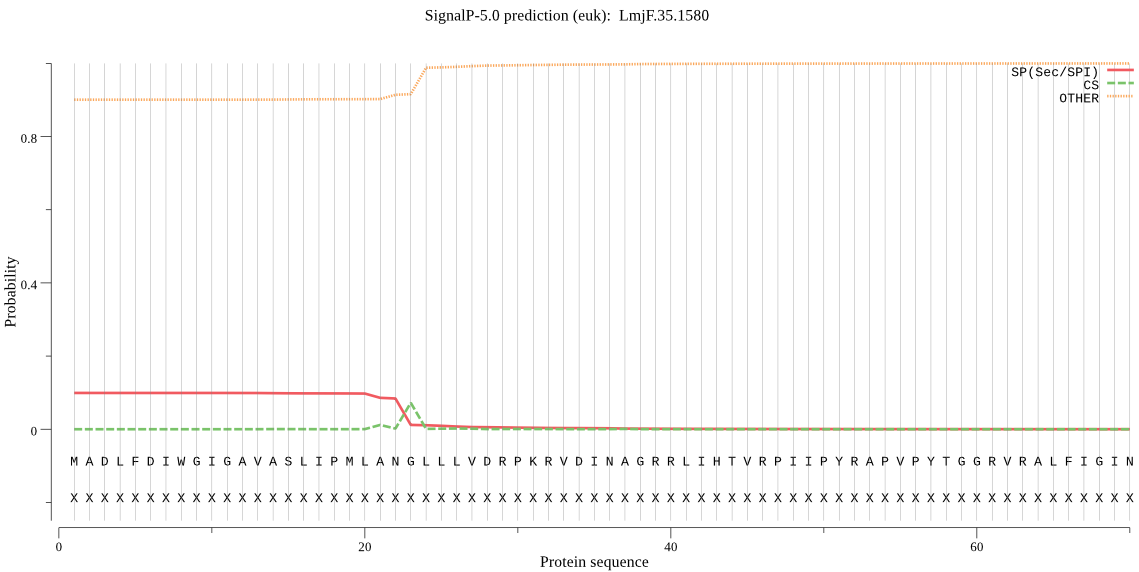
MADLFDIWGIGAVASLIPMLANGLLLVDRPKRVDINAGRRLIHTVRPIIPYRAPVPYTGG RVRALFIGINYTGMRNALRGCVNDVSSMLGTLQQISFPISECCILVDDPSFPGFCGMPTR DNIIKHMLWLTGDVRPGDVLFFHFSGHGGQAKATRDSEEKYDQCLIPLDHVKNGSILDDD LFLMLVAPLPSGVRMTCVFDCCHSASMLDLPFSYVAPRVGGGGACEYMQQVRRGNFSNGD VVMFSGCTDRGTSADVQNGGHANGAATLAFTWSLLNTHGLSYLNILLKTREELRKKGRVQ VPQLTSSKPIDLYKPFSLFGMITVNASMMHCVPQQYQQRPQSLPPQVMPPATGYPVHVPP PPQGYYPPPQSPGWGLGYPVQGIPVQQATLGVSRCPPSQYLPAPPPAVYAPPPPGQRGPP QPPPAQYTFSPLPPG
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LmjF.35.1580 | 157 S | ATRDSEEKY | 0.997 | unsp |