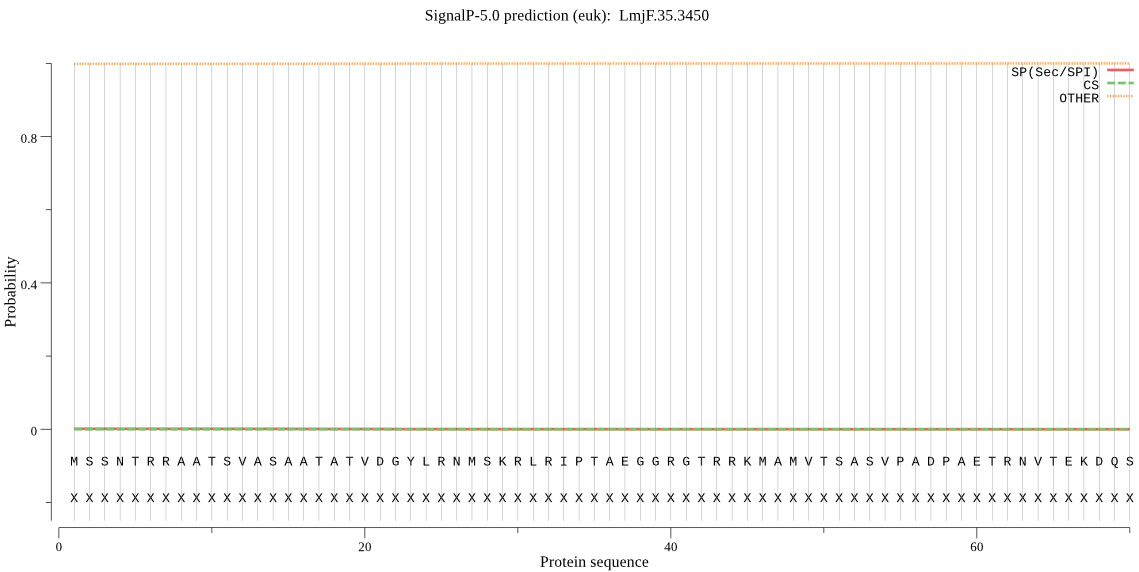
Fasta :-
MSSNTRRAATSVASAATATVDGYLRNMSKRLRIPTAEGGRGTRRKMAMVTSASVPADPAE
TRNVTEKDQSNLAADVDEWDEVELPVAFSPAKKQEESESGSLVKKGLTDDGEGEAVKHET
AYAVSTVKTEVTTTTTGEADTVTSGGVKEELVSVDTEAAASDRPLMEWAQLPLVGRPRAS
DTWRQRDPAYEAMMEQRDLLAAKRRSERIQRVCESLFALLAVLIRARLLWRESCHPGFVK
QLLRLHVSRGRGPKRSRCEDPSGKATSAYVFLKAVATAKVLVAESTKPSHRKPSLAPAWV
TCAKDAVQNKTSAAVKVLIETINDYFKLEALAAPASSSGREAPVGAAAYTVAPSSAEQDF
SDWSMPVKPGFLFTKTAERHCRVSADAPLELPHPLYFSLVFLALARVAGLSCRLVVAKLG
KTQLEKARAAGPAGDTANGYNDTSEGKDNTADGESEDSTRRPLRVLSIFEGREKRKASDR
TDASGVARAGKGSKRQRPSAGLAEDTKDREELKSKKLPTSCYWVEVWSAERESFLSVNPC
QSCATLWGASYTLSVSGHVAVDATPRYISKYSTAYAYGRRLGTCRQHRFLWRDELAWDDT
RELSEVLRATFNVAAPHTSSLAQRQQQRESRQLHSLMYSEAVPTTLNALHRHPLYVTDSD
LARHEGVYPKDANTTVGSVKGRLVYKRSAVVSLRSRDGWLREGRSLLTEDQPAYKVVAPP
ASRPFAAPSTLYGRWQTQPFEPLPLTAGDPPSIPHHGRTSWYILLDKAPPQGIVHLTQPQ
ISRVARRMKLDFCLAVVGFERRRTDEHRRGHWETVINGIVVKETDSVALLHAYEEWVQLV
QEQEATKRRQRAFHWWLLLAQRLLALKRLQDQYAKGLGAGSMPMQ