

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmjF.35.4850 | OTHER | 0.999803 | 0.000130 | 0.000067 |
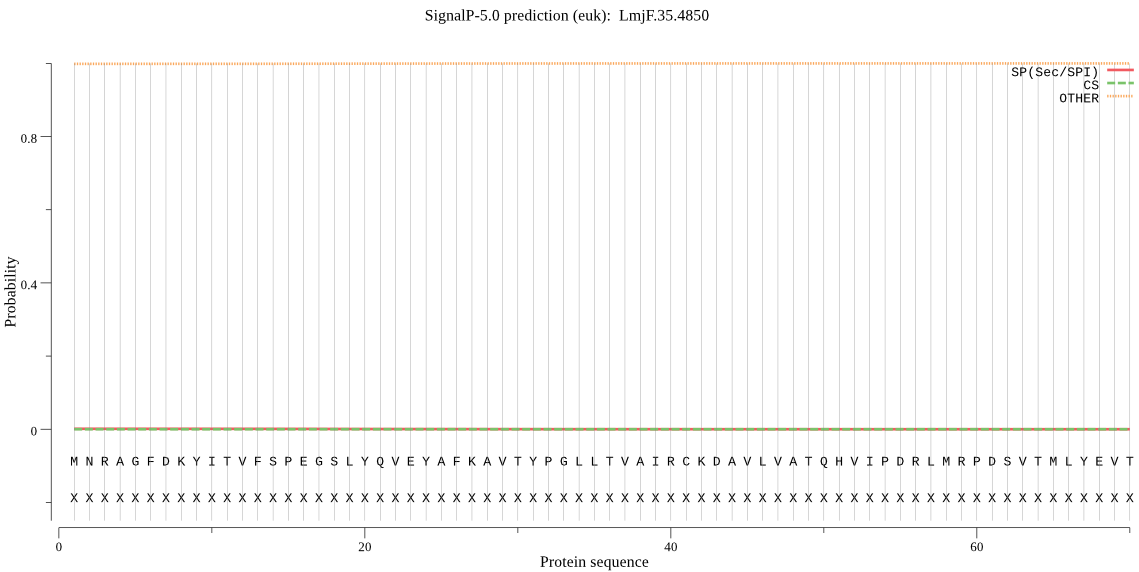
MNRAGFDKYITVFSPEGSLYQVEYAFKAVTYPGLLTVAIRCKDAVLVATQHVIPDRLMRP DSVTMLYEVTPGIGCCMTGRAPDGRALVQRAREEASDYHYRYGVQIPIAVLAKRMGDKAQ VRTQQAGLRPMGVVSTFIGMDQSDQDGALKPQIYTVDPAGWTCGHIACAVGKKQVEAMAF LEKRQKSTEFAELTQKEAAMIALAALQSAIGTAVKAKEVEVGRCTAANPAFQRVPNSEVE EWLTAVAEAD
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| LmjF.35.4850 | 187 S | KRQKSTEFA | 0.998 | unsp | LmjF.35.4850 | 194 T | FAELTQKEA | 0.99 | unsp |