

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmjF.36.0200 | SP | 0.490093 | 0.499518 | 0.010390 | CS pos: 35-36. VSC-AV. Pr: 0.7114 |
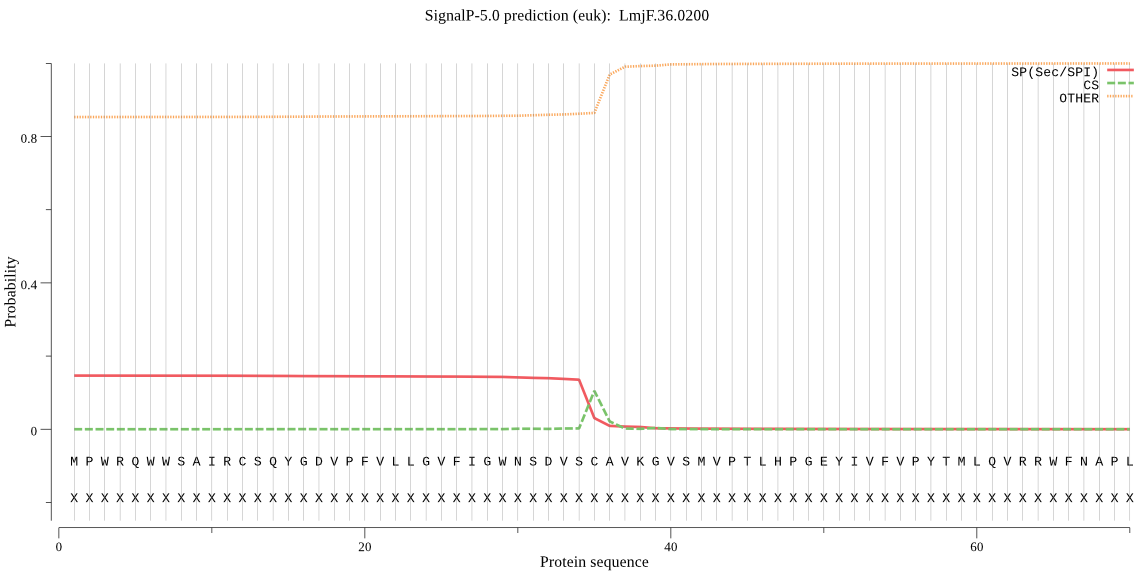
MPWRQWWSAIRCSQYGDVPFVLLGVFIGWNSDVSCAVKGVSMVPTLHPGEYIVFVPYTML QVRRWFNAPLVNLSDVVVVKVSDDLSVCKRVVKCTSSRTQAEEWGKEHYVEVMPALYSPP AAQHTNGDDEDSTEIDSVANSERAYFDYVARNTVRSKDWDSCIDRIPNPSQWVWLEGDNK SESFDSRRCGPVPIECIRGLVLASIWPSPHTLQRPPPPPRASQLH