

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| LmjF.36.1600 | OTHER | 0.999885 | 0.000028 | 0.000087 |
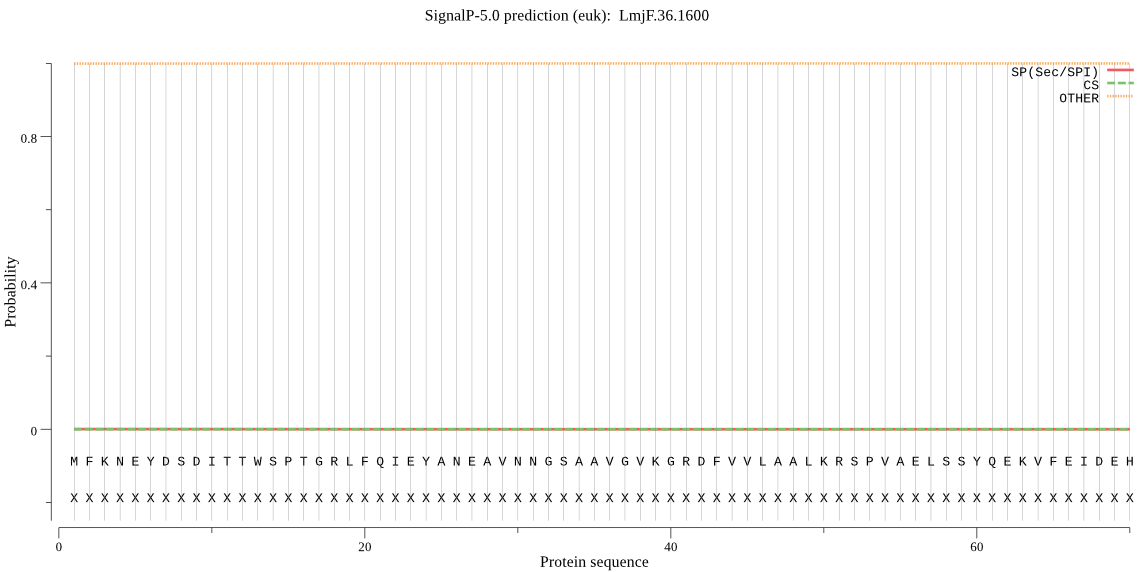
MFKNEYDSDITTWSPTGRLFQIEYANEAVNNGSAAVGVKGRDFVVLAALKRSPVAELSSY QEKVFEIDEHVGMSISGLVADGRVLARFLRTECMNYRYMYSHGMPMNQMADMIGEKHQRH IQFSGKRPFGVGLLLAGYDRQGPHLYQTVPSGDVYDYKATAMGVRSQASRTYLEKHFERF TDCTLDELVTHALKALASATSEGVELNVKNTTIAIVGKDTPFTIFEEESARRYLDGFKMR PEDRVAAADEEEVLQEQPLDVEE
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| LmjF.36.1600 | 124 S | HIQFSGKRP | 0.995 | unsp |